Alexander Zhebrak
A Prior of a Googol Gaussians: a Tensor Ring Induced Prior for Generative Models
Oct 29, 2019
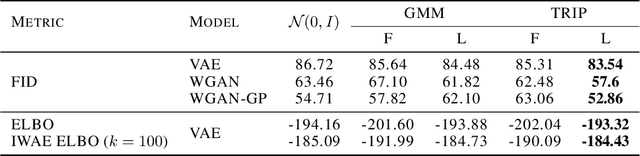


Abstract:Generative models produce realistic objects in many domains, including text, image, video, and audio synthesis. Most popular models---Generative Adversarial Networks (GANs) and Variational Autoencoders (VAEs)---usually employ a standard Gaussian distribution as a prior. Previous works show that the richer family of prior distributions may help to avoid the mode collapse problem in GANs and to improve the evidence lower bound in VAEs. We propose a new family of prior distributions---Tensor Ring Induced Prior (TRIP)---that packs an exponential number of Gaussians into a high-dimensional lattice with a relatively small number of parameters. We show that these priors improve Fr\'echet Inception Distance for GANs and Evidence Lower Bound for VAEs. We also study generative models with TRIP in the conditional generation setup with missing conditions. Altogether, we propose a novel plug-and-play framework for generative models that can be utilized in any GAN and VAE-like architectures.
Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models
Nov 29, 2018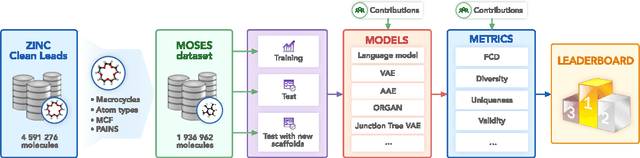
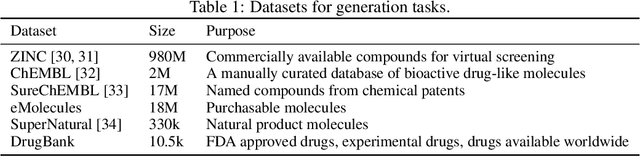

Abstract:Deep generative models such as generative adversarial networks, variational autoencoders, and autoregressive models are rapidly growing in popularity for the discovery of new molecules and materials. In this work, we introduce MOlecular SEtS (MOSES), a benchmarking platform to support research on machine learning for drug discovery. MOSES implements several popular molecular generation models and includes a set of metrics that evaluate the diversity and quality of generated molecules. MOSES is meant to standardize the research on the molecular generation and facilitate the sharing and comparison of new models. Additionally, we provide a large-scale comparison of existing state of the art models and elaborate on current challenges for generative models that might prove fertile ground for new research. Our platform and source code are freely available at https://github.com/molecularsets/
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge