Zulfat Miftahutdinov
nach0-pc: Multi-task Language Model with Molecular Point Cloud Encoder
Oct 11, 2024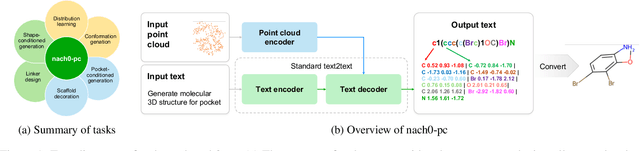
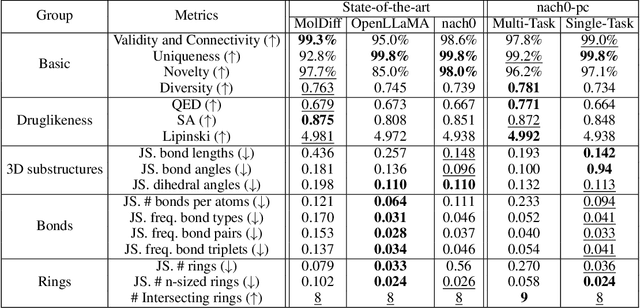
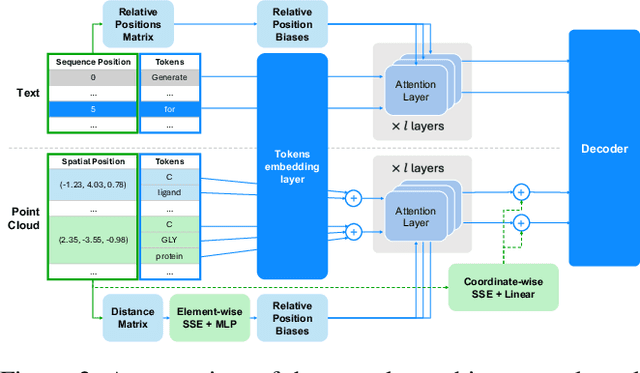
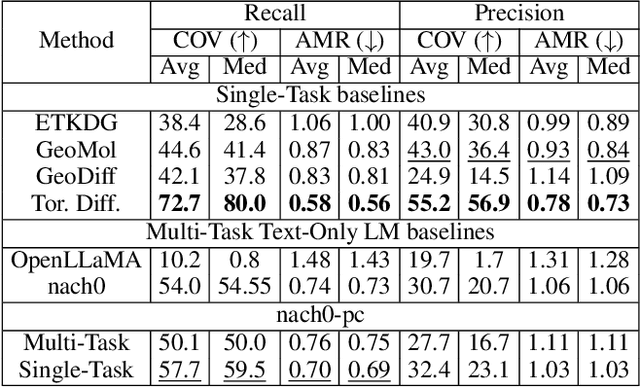
Abstract:Recent advancements have integrated Language Models (LMs) into a drug discovery pipeline. However, existing models mostly work with SMILES and SELFIES chemical string representations, which lack spatial features vital for drug discovery. Additionally, attempts to translate chemical 3D structures into text format encounter issues such as excessive length and insufficient atom connectivity information. To address these issues, we introduce nach0-pc, a model combining domain-specific encoder and textual representation to handle spatial arrangement of atoms effectively. Our approach utilizes a molecular point cloud encoder for concise and order-invariant structure representation. We introduce a novel pre-training scheme for molecular point clouds to distillate the knowledge from spatial molecular structures datasets. After fine-tuning within both single-task and multi-task frameworks, nach0-pc demonstrates performance comparable with other diffusion models in terms of generated samples quality across several established spatial molecular generation tasks. Notably, our model is a multi-task approach, in contrast to diffusion models being limited to single tasks. Additionally, it is capable of processing point cloud-related data, which language models are not capable of handling due to memory limitations. These lead to our model having reduced training and inference time while maintaining on par performance.
nach0: Multimodal Natural and Chemical Languages Foundation Model
Nov 21, 2023



Abstract:Large Language Models (LLMs) have substantially driven scientific progress in various domains, and many papers have demonstrated their ability to tackle complex problems with creative solutions. Our paper introduces a new foundation model, nach0, capable of solving various chemical and biological tasks: biomedical question answering, named entity recognition, molecular generation, molecular synthesis, attributes prediction, and others. nach0 is a multi-domain and multi-task encoder-decoder LLM pre-trained on unlabeled text from scientific literature, patents, and molecule strings to incorporate a range of chemical and linguistic knowledge. We employed instruction tuning, where specific task-related instructions are utilized to fine-tune nach0 for the final set of tasks. To train nach0 effectively, we leverage the NeMo framework, enabling efficient parallel optimization of both base and large model versions. Extensive experiments demonstrate that our model outperforms state-of-the-art baselines on single-domain and cross-domain tasks. Furthermore, it can generate high-quality outputs in molecular and textual formats, showcasing its effectiveness in multi-domain setups.
Drug and Disease Interpretation Learning with Biomedical Entity Representation Transformer
Jan 22, 2021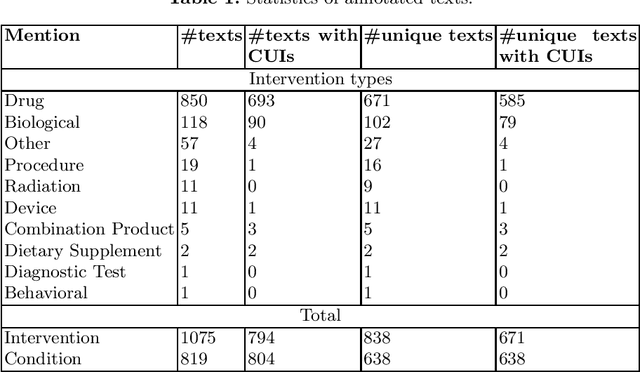
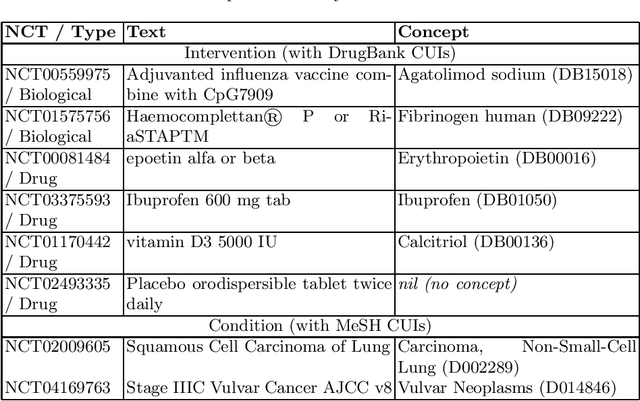
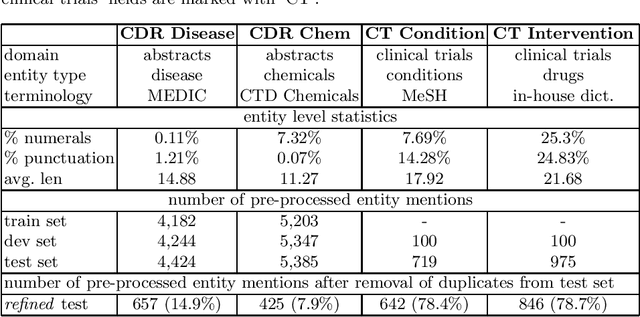
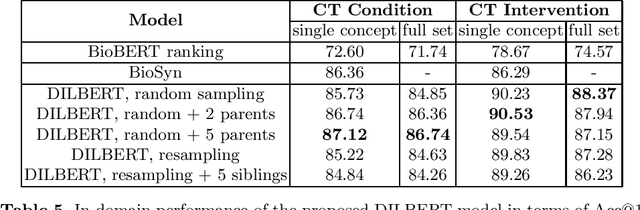
Abstract:Concept normalization in free-form texts is a crucial step in every text-mining pipeline. Neural architectures based on Bidirectional Encoder Representations from Transformers (BERT) have achieved state-of-the-art results in the biomedical domain. In the context of drug discovery and development, clinical trials are necessary to establish the efficacy and safety of drugs. We investigate the effectiveness of transferring concept normalization from the general biomedical domain to the clinical trials domain in a zero-shot setting with an absence of labeled data. We propose a simple and effective two-stage neural approach based on fine-tuned BERT architectures. In the first stage, we train a metric learning model that optimizes relative similarity of mentions and concepts via triplet loss. The model is trained on available labeled corpora of scientific abstracts to obtain vector embeddings of concept names and entity mentions from texts. In the second stage, we find the closest concept name representation in an embedding space to a given clinical mention. We evaluated several models, including state-of-the-art architectures, on a dataset of abstracts and a real-world dataset of trial records with interventions and conditions mapped to drug and disease terminologies. Extensive experiments validate the effectiveness of our approach in knowledge transfer from the scientific literature to clinical trials.
The Russian Drug Reaction Corpus and Neural Models for Drug Reactions and Effectiveness Detection in User Reviews
Apr 07, 2020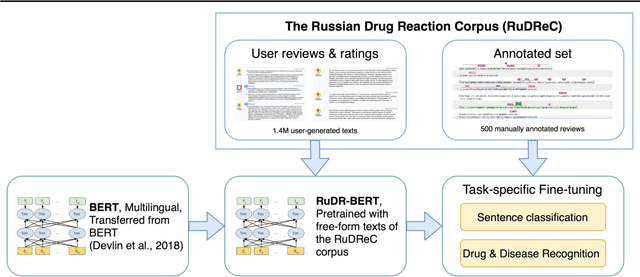
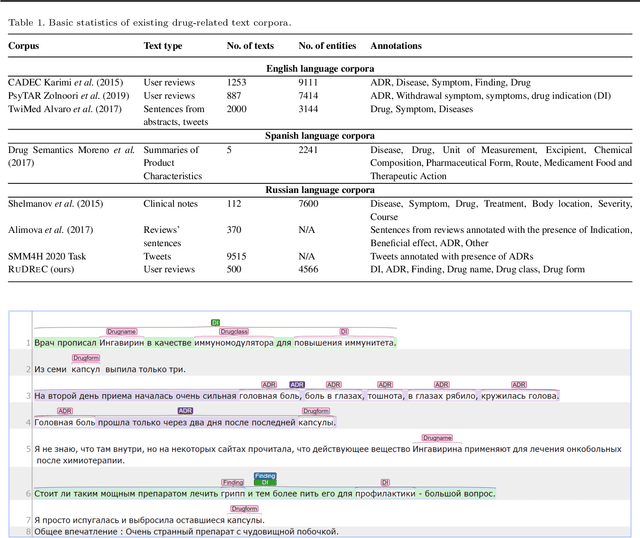
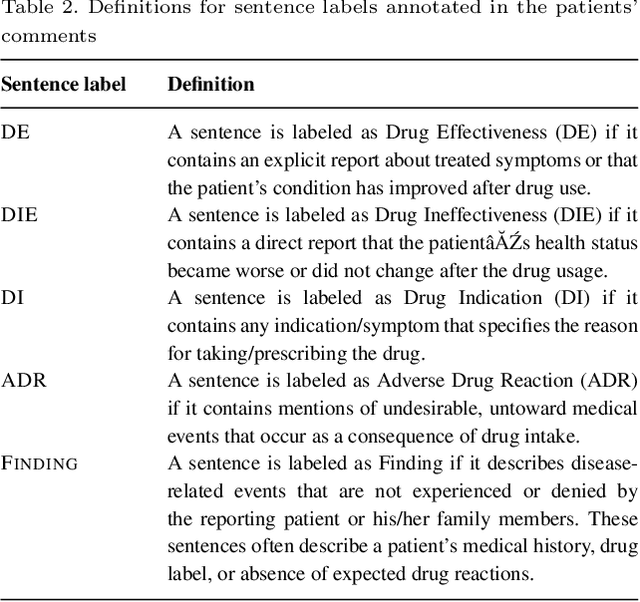
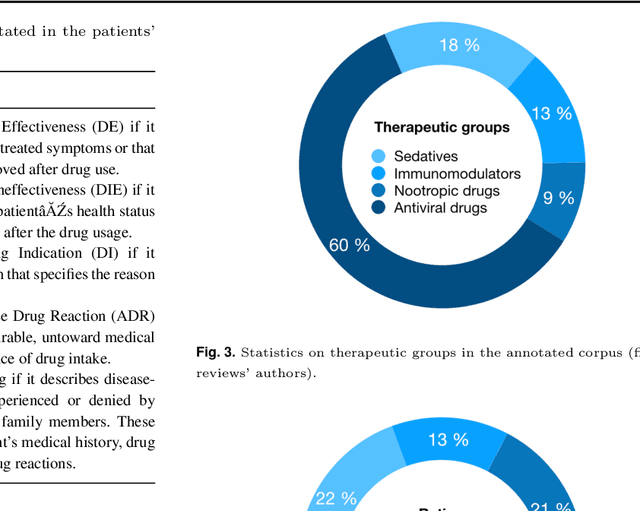
Abstract:The Russian Drug Reaction Corpus (RuDReC) is a new partially annotated corpus of consumer reviews in Russian about pharmaceutical products for the detection of health-related named entities and the effectiveness of pharmaceutical products. The corpus itself consists of two parts, the raw one and the labelled one. The raw part includes 1.4 million health-related user-generated texts collected from various Internet sources, including social media. The labelled part contains 500 consumer reviews about drug therapy with drug- and disease-related information. Labels for sentences include health-related issues or their absence. The sentences with one are additionally labelled at the expression level for identification of fine-grained subtypes such as drug classes and drug forms, drug indications, and drug reactions. Further, we present a baseline model for named entity recognition (NER) and multi-label sentence classification tasks on this corpus. The macro F1 score of 74.85% in the NER task was achieved by our RuDR-BERT model. For the sentence classification task, our model achieves the macro F1 score of 68.82% gaining 7.47% over the score of BERT model trained on Russian data. We make the RuDReC corpus and pretrained weights of domain-specific BERT models freely available at https://github.com/cimm-kzn/RuDReC
CommentsRadar: Dive into Unique Data on All Comments on the Web
Aug 16, 2019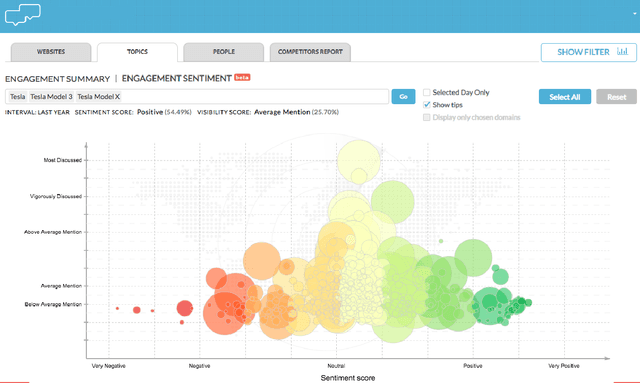
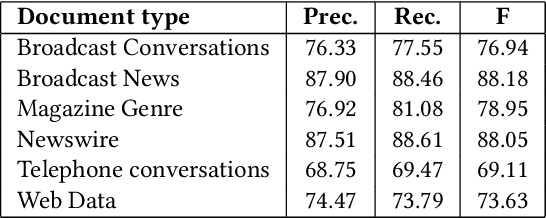
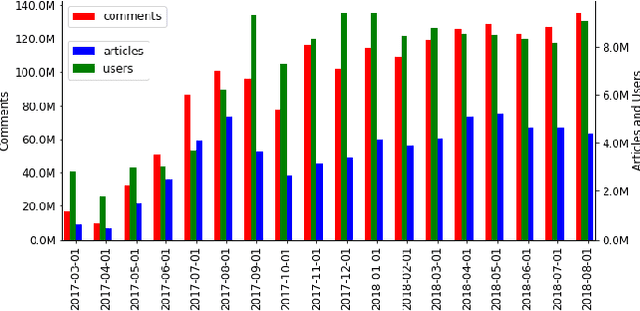
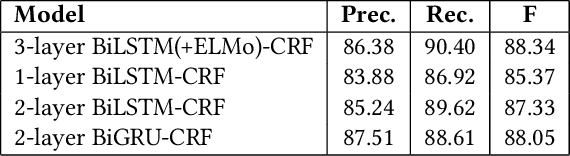
Abstract:We introduce an entity-centric search engineCommentsRadarthatpairs entity queries with articles and user opinions covering a widerange of topics from top commented sites. The engine aggregatesarticles and comments for these articles, extracts named entities,links them together and with knowledge base entries, performssentiment analysis, and aggregates the results, aiming to mine fortemporal trends and other insights. In this work, we present thegeneral engine, discuss the models used for all steps of this pipeline,and introduce several case studies that discover important insightsfrom online commenting data.
Deep Neural Models for Medical Concept Normalization in User-Generated Texts
Jul 18, 2019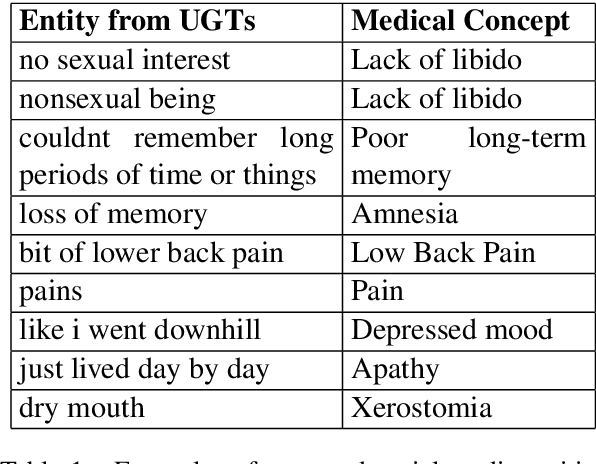

Abstract:In this work, we consider the medical concept normalization problem, i.e., the problem of mapping a health-related entity mention in a free-form text to a concept in a controlled vocabulary, usually to the standard thesaurus in the Unified Medical Language System (UMLS). This is a challenging task since medical terminology is very different when coming from health care professionals or from the general public in the form of social media texts. We approach it as a sequence learning problem with powerful neural networks such as recurrent neural networks and contextualized word representation models trained to obtain semantic representations of social media expressions. Our experimental evaluation over three different benchmarks shows that neural architectures leverage the semantic meaning of the entity mention and significantly outperform an existing state of the art models.
Sequence Learning with RNNs for Medical Concept Normalization in User-Generated Texts
Nov 29, 2018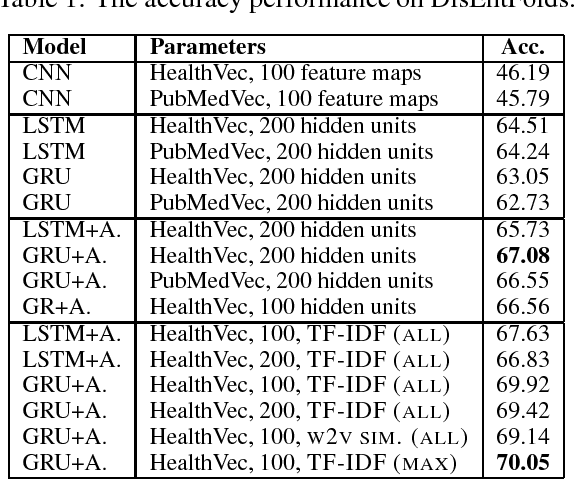

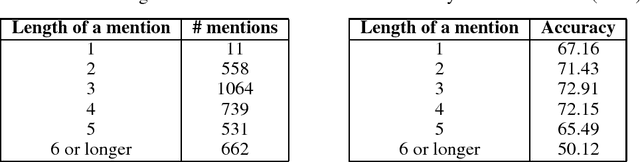
Abstract:In this work, we consider the medical concept normalization problem, i.e., the problem of mapping a disease mention in free-form text to a concept in a controlled vocabulary, usually to the standard thesaurus in the Unified Medical Language System (UMLS). This task is challenging since medical terminology is very different when coming from health care professionals or from the general public in the form of social media texts. We approach it as a sequence learning problem, with recurrent neural networks trained to obtain semantic representations of one- and multi-word expressions. We develop end-to-end neural architectures tailored specifically to medical concept normalization, including bidirectional LSTM and GRU with an attention mechanism and additional semantic similarity features based on UMLS. Our evaluation over a standard benchmark shows that our model improves over a state of the art baseline for classification based on CNNs.
* Machine Learning for Health (ML4H) Workshop at NeurIPS 2018 arXiv:1811.07216
An Encoder-Decoder Model for ICD-10 Coding of Death Certificates
Dec 04, 2017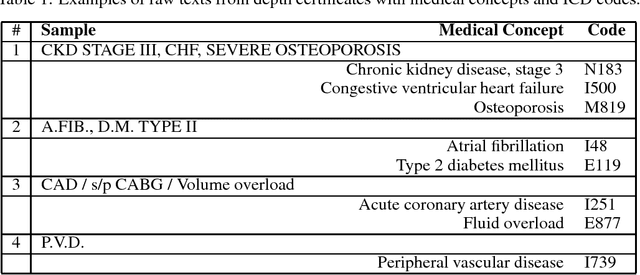
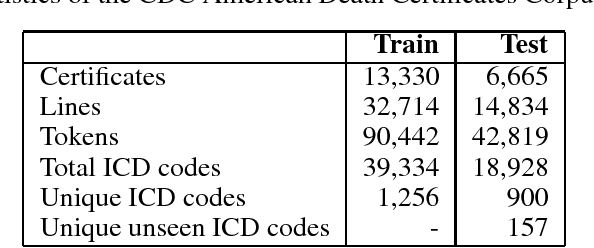
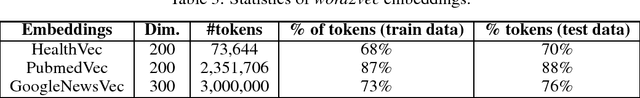
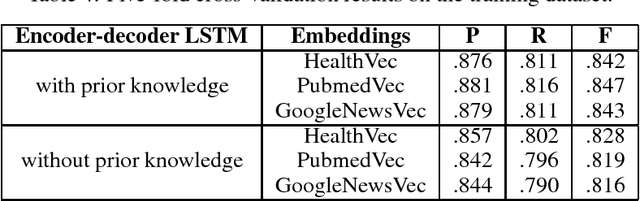
Abstract:Information extraction from textual documents such as hospital records and healthrelated user discussions has become a topic of intense interest. The task of medical concept coding is to map a variable length text to medical concepts and corresponding classification codes in some external system or ontology. In this work, we utilize recurrent neural networks to automatically assign ICD-10 codes to fragments of death certificates written in English. We develop end-to-end neural architectures directly tailored to the task, including basic encoder-decoder architecture for statistical translation. In order to incorporate prior knowledge, we concatenate cosine similarities vector among the text and dictionary entry to the encoded state. Being applied to a standard benchmark from CLEF eHealth 2017 challenge, our model achieved F-measure of 85.01% on a full test set with significant improvement as compared to the average score of 62.2% for all official participants approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge