nach0-pc: Multi-task Language Model with Molecular Point Cloud Encoder
Paper and Code
Oct 11, 2024
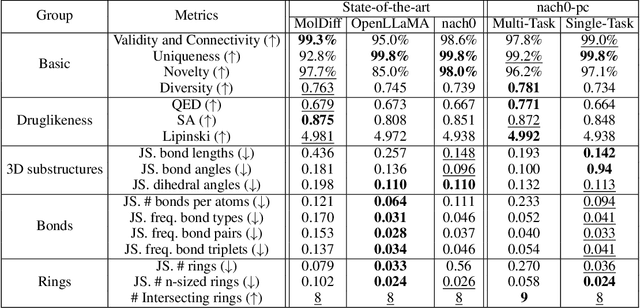
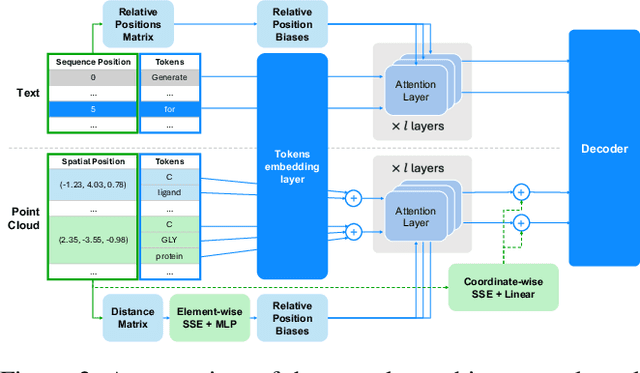
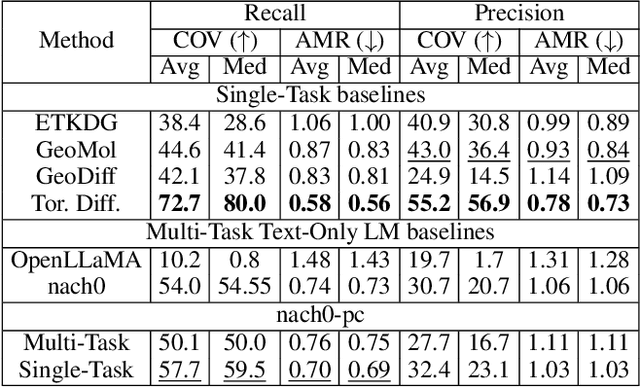
Recent advancements have integrated Language Models (LMs) into a drug discovery pipeline. However, existing models mostly work with SMILES and SELFIES chemical string representations, which lack spatial features vital for drug discovery. Additionally, attempts to translate chemical 3D structures into text format encounter issues such as excessive length and insufficient atom connectivity information. To address these issues, we introduce nach0-pc, a model combining domain-specific encoder and textual representation to handle spatial arrangement of atoms effectively. Our approach utilizes a molecular point cloud encoder for concise and order-invariant structure representation. We introduce a novel pre-training scheme for molecular point clouds to distillate the knowledge from spatial molecular structures datasets. After fine-tuning within both single-task and multi-task frameworks, nach0-pc demonstrates performance comparable with other diffusion models in terms of generated samples quality across several established spatial molecular generation tasks. Notably, our model is a multi-task approach, in contrast to diffusion models being limited to single tasks. Additionally, it is capable of processing point cloud-related data, which language models are not capable of handling due to memory limitations. These lead to our model having reduced training and inference time while maintaining on par performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge