Pierangela Bruno
Improving ASP-based ORS Schedules through Machine Learning Predictions
Jul 22, 2025Abstract:The Operating Room Scheduling (ORS) problem deals with the optimization of daily operating room surgery schedules. It is a challenging problem subject to many constraints, like to determine the starting time of different surgeries and allocating the required resources, including the availability of beds in different department units. Recently, solutions to this problem based on Answer Set Programming (ASP) have been delivered. Such solutions are overall satisfying but, when applied to real data, they can currently only verify whether the encoding aligns with the actual data and, at most, suggest alternative schedules that could have been computed. As a consequence, it is not currently possible to generate provisional schedules. Furthermore, the resulting schedules are not always robust. In this paper, we integrate inductive and deductive techniques for solving these issues. We first employ machine learning algorithms to predict the surgery duration, from historical data, to compute provisional schedules. Then, we consider the confidence of such predictions as an additional input to our problem and update the encoding correspondingly in order to compute more robust schedules. Results on historical data from the ASL1 Liguria in Italy confirm the viability of our integration. Under consideration in Theory and Practice of Logic Programming (TPLP).
μ-Net: A Deep Learning-Based Architecture for μ-CT Segmentation
Jun 24, 2024


Abstract:X-ray computed microtomography ({\mu}-CT) is a non-destructive technique that can generate high-resolution 3D images of the internal anatomy of medical and biological samples. These images enable clinicians to examine internal anatomy and gain insights into the disease or anatomical morphology. However, extracting relevant information from 3D images requires semantic segmentation of the regions of interest, which is usually done manually and results time-consuming and tedious. In this work, we propose a novel framework that uses a convolutional neural network (CNN) to automatically segment the full morphology of the heart of Carassius auratus. The framework employs an optimized 2D CNN architecture that can infer a 3D segmentation of the sample, avoiding the high computational cost of a 3D CNN architecture. We tackle the challenges of handling large and high-resoluted image data (over a thousand pixels in each dimension) and a small training database (only three samples) by proposing a standard protocol for data normalization and processing. Moreover, we investigate how the noise, contrast, and spatial resolution of the sample and the training of the architecture are affected by the reconstruction technique, which depends on the number of input images. Experiments show that our framework significantly reduces the time required to segment new samples, allowing a faster microtomography analysis of the Carassius auratus heart shape. Furthermore, our framework can work with any bio-image (biological and medical) from {\mu}-CT with high-resolution and small dataset size
Data Augmentation: a Combined Inductive-Deductive Approach featuring Answer Set Programming
Oct 22, 2023Abstract:Although the availability of a large amount of data is usually given for granted, there are relevant scenarios where this is not the case; for instance, in the biomedical/healthcare domain, some applications require to build huge datasets of proper images, but the acquisition of such images is often hard for different reasons (e.g., accessibility, costs, pathology-related variability), thus causing limited and usually imbalanced datasets. Hence, the need for synthesizing photo-realistic images via advanced Data Augmentation techniques is crucial. In this paper we propose a hybrid inductive-deductive approach to the problem; in particular, starting from a limited set of real labeled images, the proposed framework makes use of logic programs for declaratively specifying the structure of new images, that is guaranteed to comply with both a set of constraints coming from the domain knowledge and some specific desiderata. The resulting labeled images undergo a dedicated process based on Deep Learning in charge of creating photo-realistic images that comply with the generated label.
How can we learn from challenges? A statistical approach to driving future algorithm development
Jun 17, 2021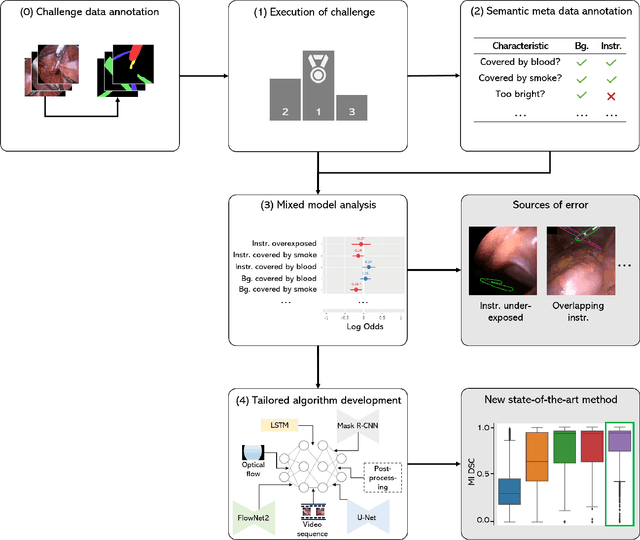
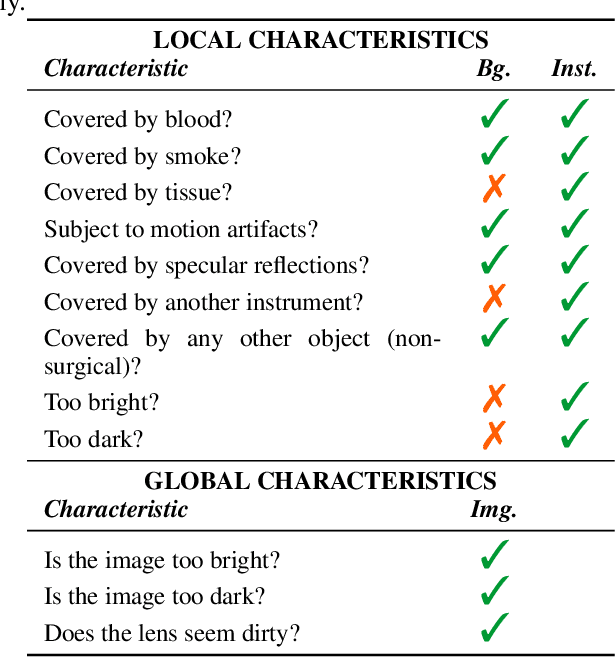
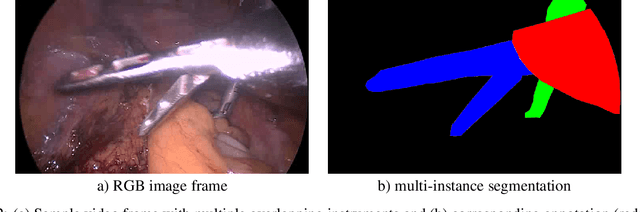
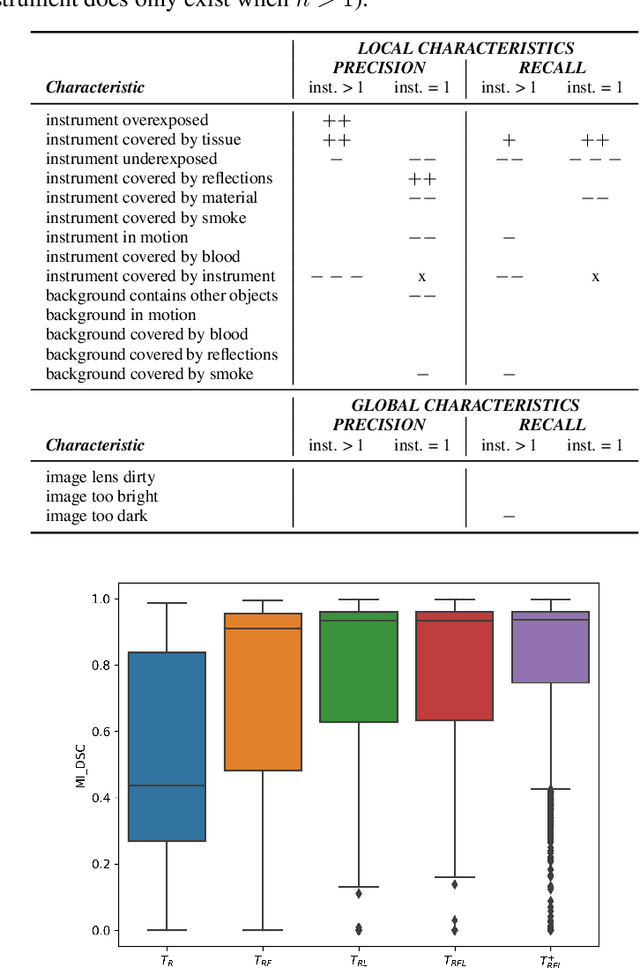
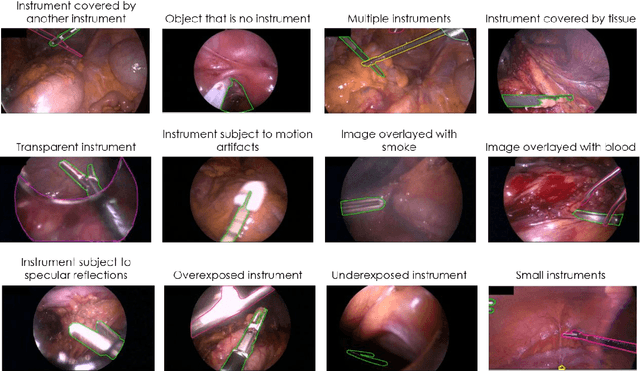
Abstract:Challenges have become the state-of-the-art approach to benchmark image analysis algorithms in a comparative manner. While the validation on identical data sets was a great step forward, results analysis is often restricted to pure ranking tables, leaving relevant questions unanswered. Specifically, little effort has been put into the systematic investigation on what characterizes images in which state-of-the-art algorithms fail. To address this gap in the literature, we (1) present a statistical framework for learning from challenges and (2) instantiate it for the specific task of instrument instance segmentation in laparoscopic videos. Our framework relies on the semantic meta data annotation of images, which serves as foundation for a General Linear Mixed Models (GLMM) analysis. Based on 51,542 meta data annotations performed on 2,728 images, we applied our approach to the results of the Robust Medical Instrument Segmentation Challenge (ROBUST-MIS) challenge 2019 and revealed underexposure, motion and occlusion of instruments as well as the presence of smoke or other objects in the background as major sources of algorithm failure. Our subsequent method development, tailored to the specific remaining issues, yielded a deep learning model with state-of-the-art overall performance and specific strengths in the processing of images in which previous methods tended to fail. Due to the objectivity and generic applicability of our approach, it could become a valuable tool for validation in the field of medical image analysis and beyond. and segmentation of small, crossing, moving and transparent instrument(s) (parts).
Heidelberg Colorectal Data Set for Surgical Data Science in the Sensor Operating Room
May 28, 2020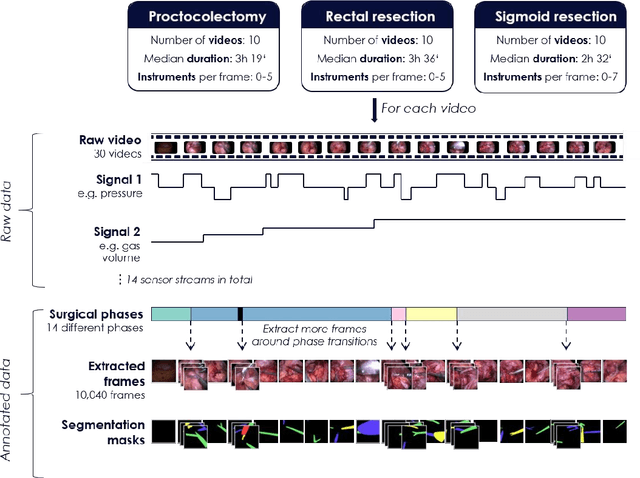
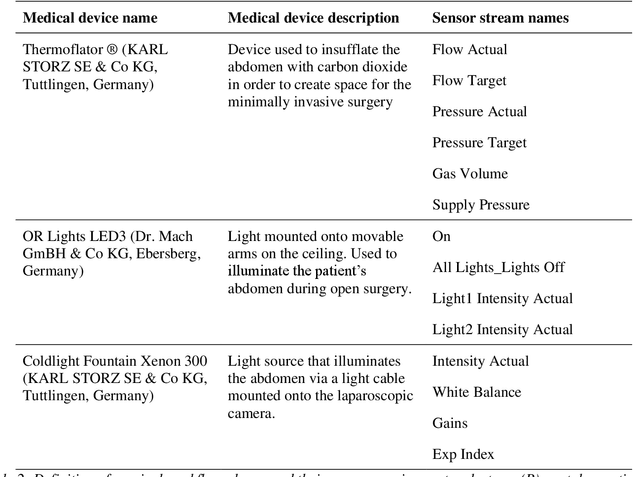
Abstract:Image-based tracking of medical instruments is an integral part of many surgical data science applications. Previous research has addressed the tasks of detecting, segmenting and tracking medical instruments based on laparoscopic video data. However, the methods proposed still tend to fail when applied to challenging images and do not generalize well to data they have not been trained on. This paper introduces the Heidelberg Colorectal (HeiCo) data set - the first publicly available data set enabling comprehensive benchmarking of medical instrument detection and segmentation algorithms with a specific emphasis on robustness and generalization capabilities of the methods. Our data set comprises 30 laparoscopic videos and corresponding sensor data from medical devices in the operating room for three different types of laparoscopic surgery. Annotations include surgical phase labels for all frames in the videos as well as instance-wise segmentation masks for surgical instruments in more than 10,000 individual frames. The data has successfully been used to organize international competitions in the scope of the Endoscopic Vision Challenges (EndoVis) 2017 and 2019.
Robust Medical Instrument Segmentation Challenge 2019
Mar 23, 2020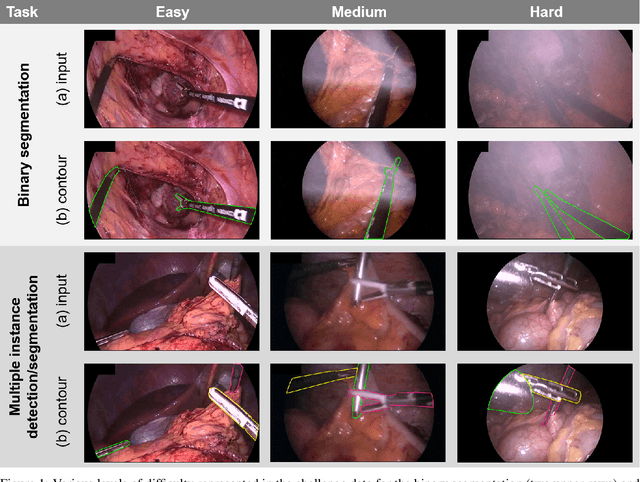
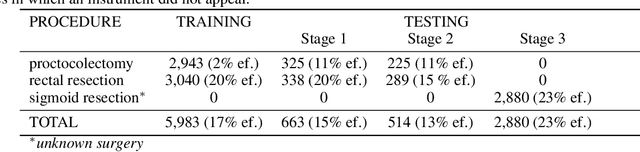
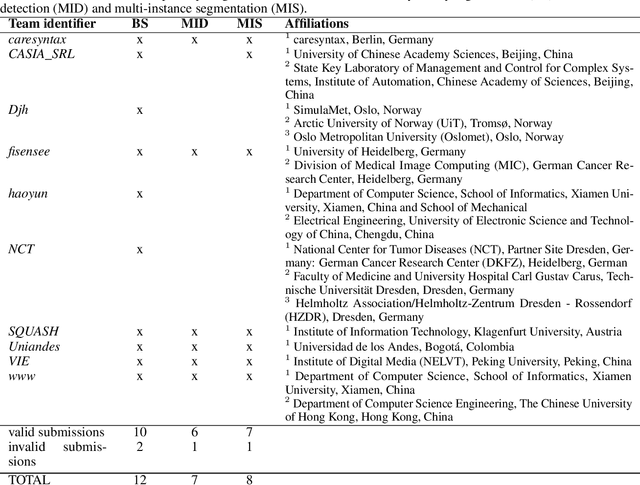
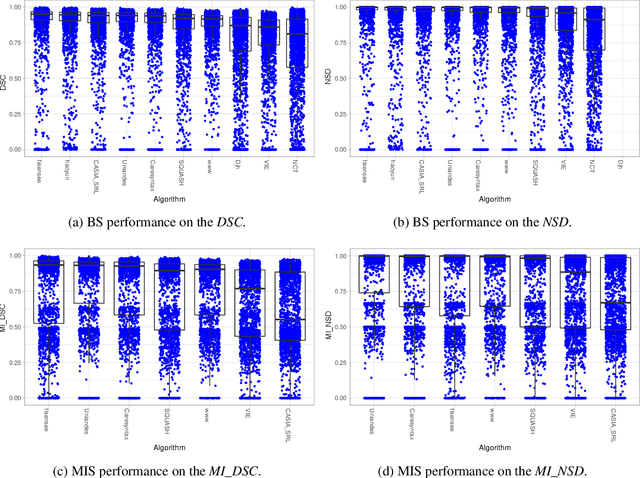
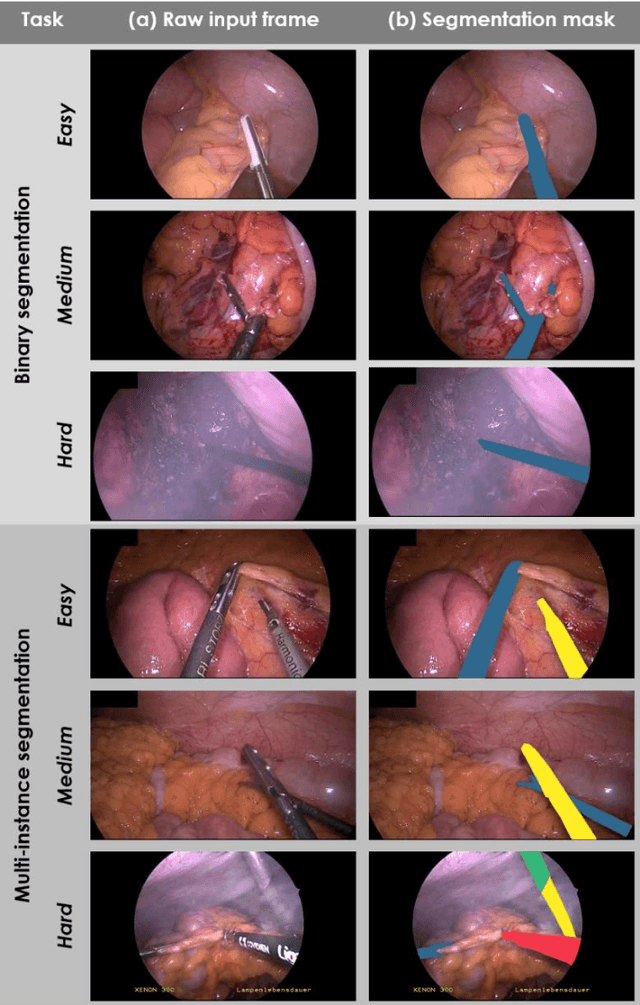
Abstract:Intraoperative tracking of laparoscopic instruments is often a prerequisite for computer and robotic-assisted interventions. While numerous methods for detecting, segmenting and tracking of medical instruments based on endoscopic video images have been proposed in the literature, key limitations remain to be addressed: Firstly, robustness, that is, the reliable performance of state-of-the-art methods when run on challenging images (e.g. in the presence of blood, smoke or motion artifacts). Secondly, generalization; algorithms trained for a specific intervention in a specific hospital should generalize to other interventions or institutions. In an effort to promote solutions for these limitations, we organized the Robust Medical Instrument Segmentation (ROBUST-MIS) challenge as an international benchmarking competition with a specific focus on the robustness and generalization capabilities of algorithms. For the first time in the field of endoscopic image processing, our challenge included a task on binary segmentation and also addressed multi-instance detection and segmentation. The challenge was based on a surgical data set comprising 10,040 annotated images acquired from a total of 30 surgical procedures from three different types of surgery. The validation of the competing methods for the three tasks (binary segmentation, multi-instance detection and multi-instance segmentation) was performed in three different stages with an increasing domain gap between the training and the test data. The results confirm the initial hypothesis, namely that algorithm performance degrades with an increasing domain gap. While the average detection and segmentation quality of the best-performing algorithms is high, future research should concentrate on detection and segmentation of small, crossing, moving and transparent instrument(s) (parts).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge