Perla Mayo
Diffusion Probabilistic Models for Compressive SAR Imaging
Apr 23, 2025Abstract:Compressed sensing Synthetic Aperture Radar (SAR) image formation, formulated as an inverse problem and solved with traditional iterative optimization methods can be very computationally expensive. We investigate the use of denoising diffusion probabilistic models for compressive SAR image reconstruction, where the diffusion model is guided by a poor initial reconstruction from sub-sampled data obtained via standard imaging methods. We present results on real SAR data and compare our compressively sampled diffusion model reconstruction with standard image reconstruction methods utilizing the full data set, demonstrating the potential performance gains in imaging quality.
Improved Patch Denoising Diffusion Probabilistic Models for Magnetic Resonance Fingerprinting
Oct 29, 2024


Abstract:Magnetic Resonance Fingerprinting (MRF) is a time-efficient approach to quantitative MRI, enabling the mapping of multiple tissue properties from a single, accelerated scan. However, achieving accurate reconstructions remains challenging, particularly in highly accelerated and undersampled acquisitions, which are crucial for reducing scan times. While deep learning techniques have advanced image reconstruction, the recent introduction of diffusion models offers new possibilities for imaging tasks, though their application in the medical field is still emerging. Notably, diffusion models have not yet been explored for the MRF problem. In this work, we propose for the first time a conditional diffusion probabilistic model for MRF image reconstruction. Qualitative and quantitative comparisons on in-vivo brain scan data demonstrate that the proposed approach can outperform established deep learning and compressed sensing algorithms for MRF reconstruction. Extensive ablation studies also explore strategies to improve computational efficiency of our approach.
StoDIP: Efficient 3D MRF image reconstruction with deep image priors and stochastic iterations
Aug 05, 2024


Abstract:Magnetic Resonance Fingerprinting (MRF) is a time-efficient approach to quantitative MRI for multiparametric tissue mapping. The reconstruction of quantitative maps requires tailored algorithms for removing aliasing artefacts from the compressed sampled MRF acquisitions. Within approaches found in the literature, many focus solely on two-dimensional (2D) image reconstruction, neglecting the extension to volumetric (3D) scans despite their higher relevance and clinical value. A reason for this is that transitioning to 3D imaging without appropriate mitigations presents significant challenges, including increased computational cost and storage requirements, and the need for large amount of ground-truth (artefact-free) data for training. To address these issues, we introduce StoDIP, a new algorithm that extends the ground-truth-free Deep Image Prior (DIP) reconstruction to 3D MRF imaging. StoDIP employs memory-efficient stochastic updates across the multicoil MRF data, a carefully selected neural network architecture, as well as faster nonuniform FFT (NUFFT) transformations. This enables a faster convergence compared against a conventional DIP implementation without these features. Tested on a dataset of whole-brain scans from healthy volunteers, StoDIP demonstrated superior performance over the ground-truth-free reconstruction baselines, both quantitatively and qualitatively.
Deep Image Priors for Magnetic Resonance Fingerprinting with pretrained Bloch-consistent denoising autoencoders
Jul 29, 2024



Abstract:The estimation of multi-parametric quantitative maps from Magnetic Resonance Fingerprinting (MRF) compressed sampled acquisitions, albeit successful, remains a challenge due to the high underspampling rate and artifacts naturally occuring during image reconstruction. Whilst state-of-the-art DL methods can successfully address the task, to fully exploit their capabilities they often require training on a paired dataset, in an area where ground truth is seldom available. In this work, we propose a method that combines a deep image prior (DIP) module that, without ground truth and in conjunction with a Bloch consistency enforcing autoencoder, can tackle the problem, resulting in a method faster and of equivalent or better accuracy than DIP-MRF.
Detection of Parasitic Eggs from Microscopy Images and the emergence of a new dataset
Mar 06, 2022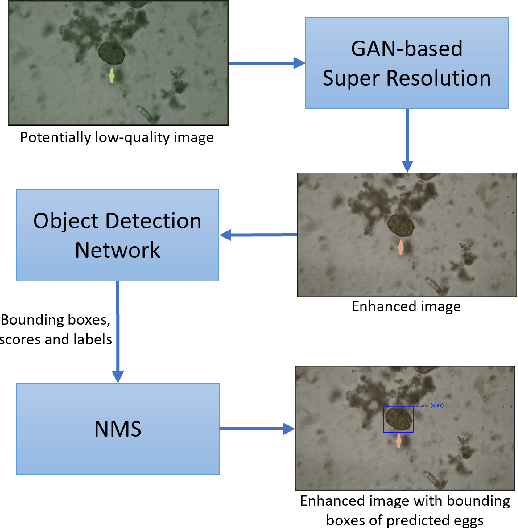
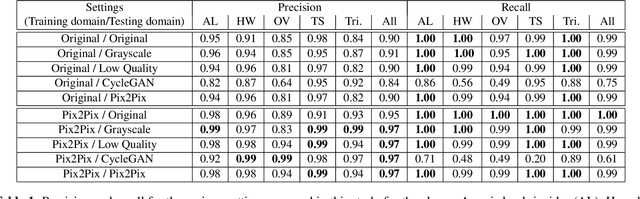
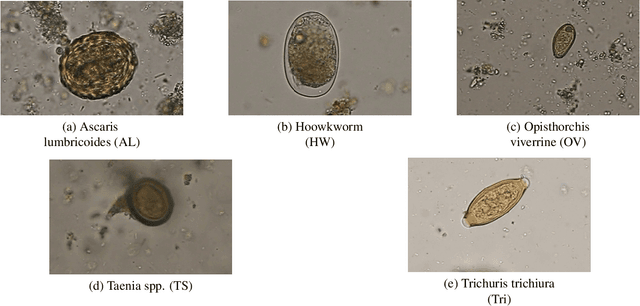
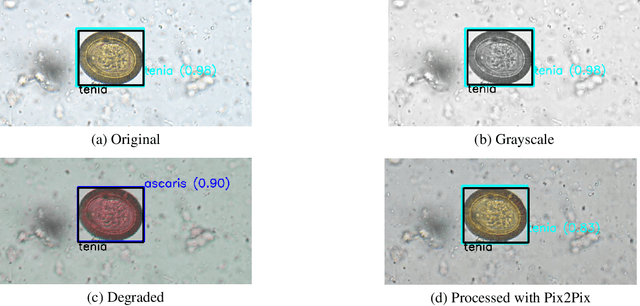
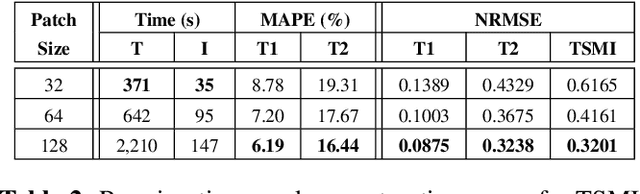
Abstract:Automatic detection of parasitic eggs in microscopy images has the potential to increase the efficiency of human experts whilst also providing an objective assessment. The time saved by such a process would both help ensure a prompt treatment to patients, and off-load excessive work from experts' shoulders. Advances in deep learning inspired us to exploit successful architectures for detection, adapting them to tackle a different domain. We propose a framework that exploits two such state-of-the-art models. Specifically, we demonstrate results produced by both a Generative Adversarial Network (GAN) and Faster-RCNN, for image enhancement and object detection respectively, on microscopy images of varying quality. The use of these techniques yields encouraging results, though further improvements are still needed for certain egg types whose detection still proves challenging. As a result, a new dataset has been created and made publicly available, providing an even wider range of classes and variability.
Representation Learning via Cauchy Convolutional Sparse Coding
Aug 08, 2020



Abstract:In representation learning, Convolutional Sparse Coding (CSC) enables unsupervised learning of features by jointly optimising both an \(\ell_2\)-norm fidelity term and a sparsity enforcing penalty. This work investigates using a regularisation term derived from an assumed Cauchy prior for the coefficients of the feature maps of a CSC generative model. The sparsity penalty term resulting from this prior is solved via its proximal operator, which is then applied iteratively, element-wise, on the coefficients of the feature maps to optimise the CSC cost function. The performance of the proposed Iterative Cauchy Thresholding (ICT) algorithm in reconstructing natural images is compared against the common choice of \(\ell_1\)-norm optimised via soft and hard thresholding. ICT outperforms IHT and IST in most of these reconstruction experiments across various datasets, with an average PSNR of up to 11.30 and 7.04 above ISTA and IHT respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge