Thanarat H. Chalidabhongse
ICIP 2022 Challenge on Parasitic Egg Detection and Classification in Microscopic Images: Dataset, Methods and Results
Aug 11, 2022
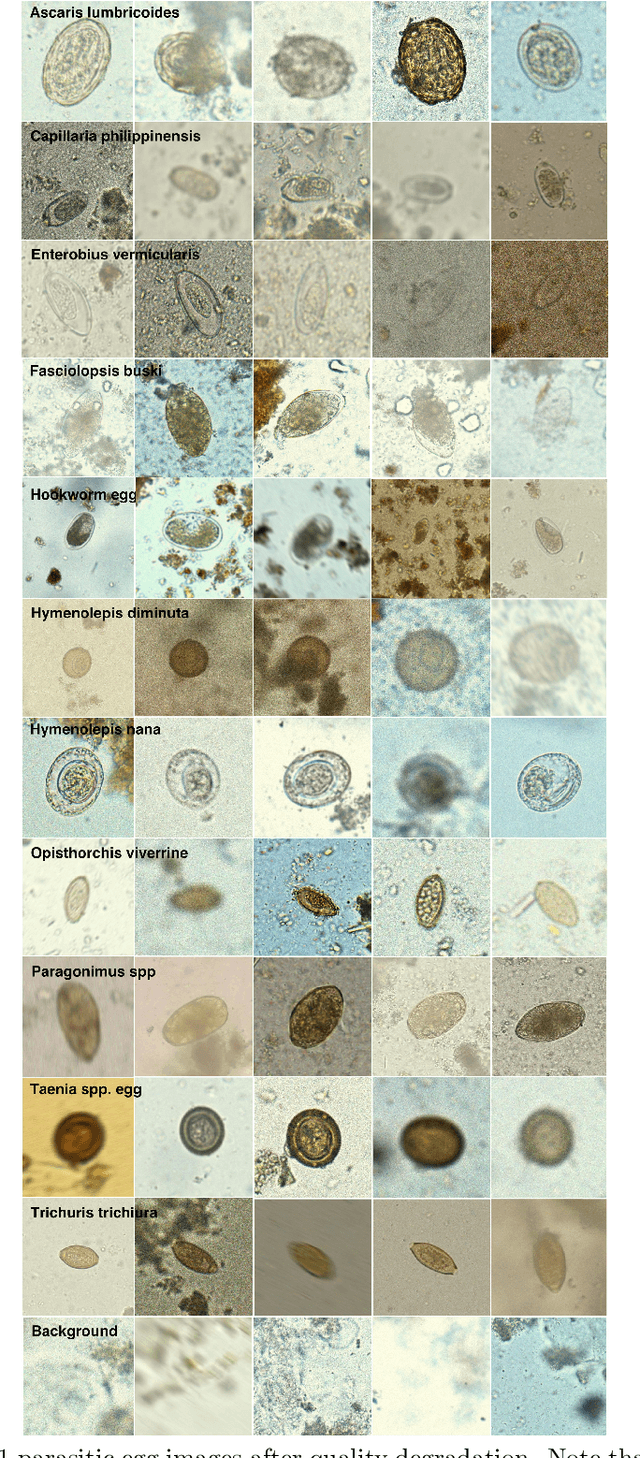
Abstract:Manual examination of faecal smear samples to identify the existence of parasitic eggs is very time-consuming and can only be done by specialists. Therefore, an automated system is required to tackle this problem since it can relate to serious intestinal parasitic infections. This paper reviews the ICIP 2022 Challenge on parasitic egg detection and classification in microscopic images. We describe a new dataset for this application, which is the largest dataset of its kind. The methods used by participants in the challenge are summarised and discussed along with their results.
Detection of Parasitic Eggs from Microscopy Images and the emergence of a new dataset
Mar 06, 2022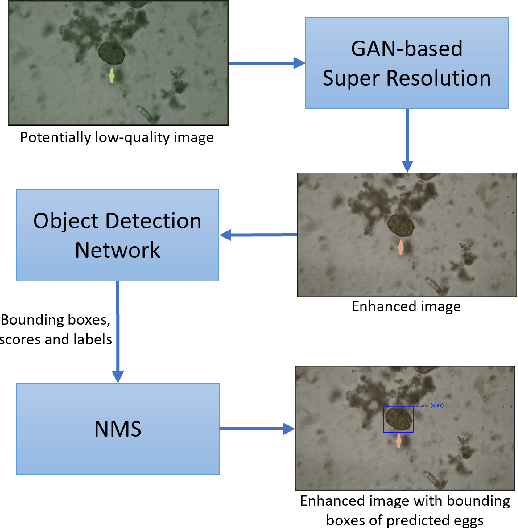
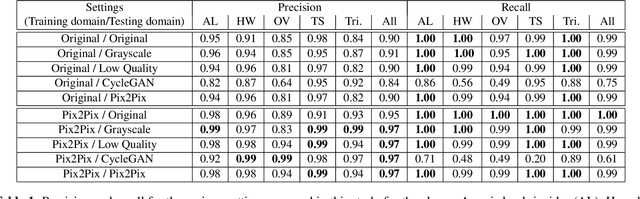
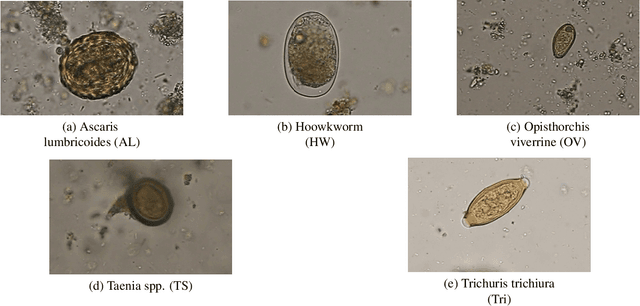
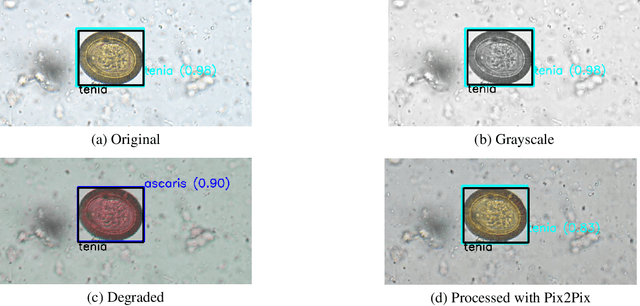
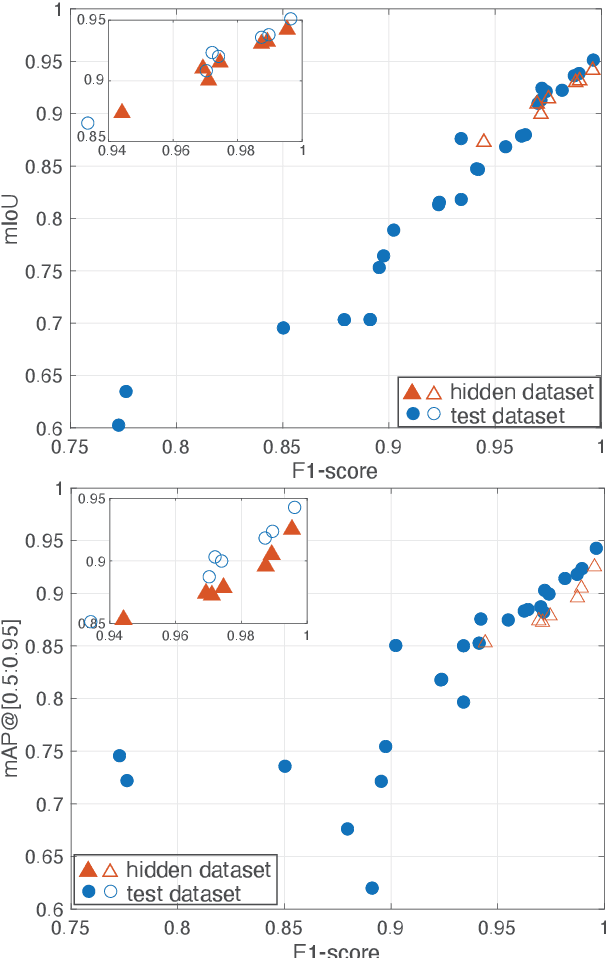
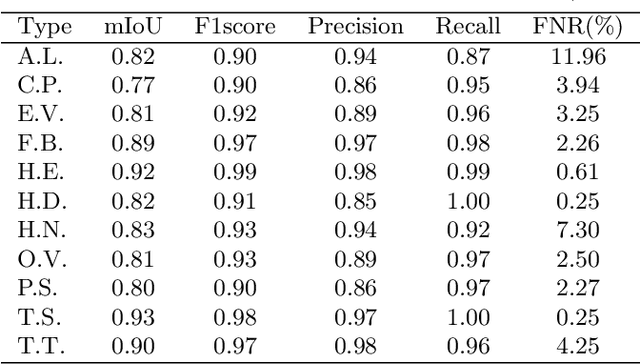
Abstract:Automatic detection of parasitic eggs in microscopy images has the potential to increase the efficiency of human experts whilst also providing an objective assessment. The time saved by such a process would both help ensure a prompt treatment to patients, and off-load excessive work from experts' shoulders. Advances in deep learning inspired us to exploit successful architectures for detection, adapting them to tackle a different domain. We propose a framework that exploits two such state-of-the-art models. Specifically, we demonstrate results produced by both a Generative Adversarial Network (GAN) and Faster-RCNN, for image enhancement and object detection respectively, on microscopy images of varying quality. The use of these techniques yields encouraging results, though further improvements are still needed for certain egg types whose detection still proves challenging. As a result, a new dataset has been created and made publicly available, providing an even wider range of classes and variability.
Parasitic Egg Detection and Classification in Low-cost Microscopic Images using Transfer Learning
Jul 02, 2021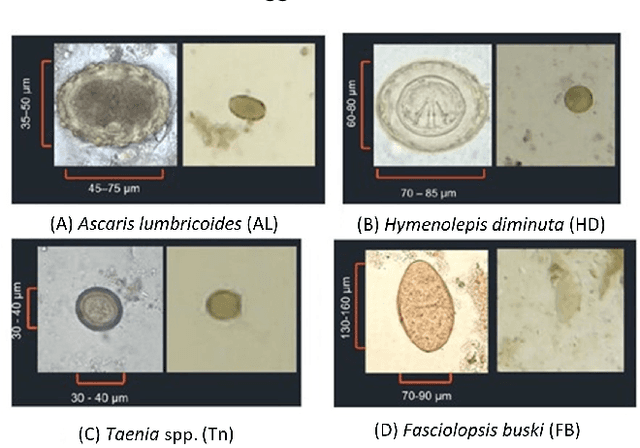
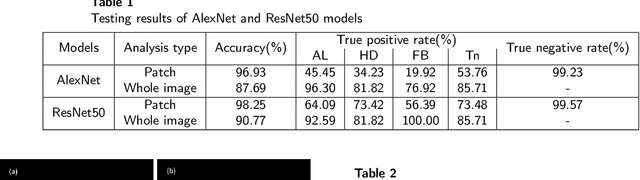
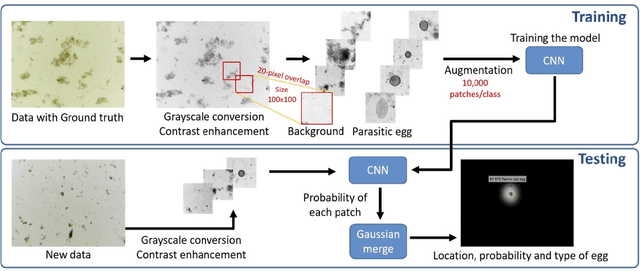
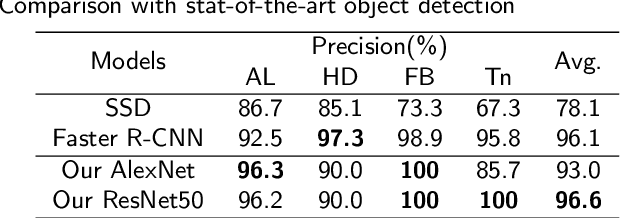
Abstract:Intestinal parasitic infection leads to several morbidities to humans worldwide, especially in tropical countries. The traditional diagnosis usually relies on manual analysis from microscopic images which is prone to human error due to morphological similarity of different parasitic eggs and abundance of impurities in a sample. Many studies have developed automatic systems for parasite egg detection to reduce human workload. However, they work with high quality microscopes, which unfortunately remain unaffordable in some rural areas. Our work thus exploits a benefit of a low-cost USB microscope. This instrument however provides poor quality of images due to limitation of magnification (10x), causing difficulty in parasite detection and species classification. In this paper, we propose a CNN-based technique using transfer learning strategy to enhance the efficiency of automatic parasite classification in poor-quality microscopic images. The patch-based technique with sliding window is employed to search for location of the eggs. Two networks, AlexNet and ResNet50, are examined with a trade-off between architecture size and classification performance. The results show that our proposed framework outperforms the state-of-the-art object recognition methods. Our system combined with final decision from an expert may improve the real faecal examination with low-cost microscopes.
Analysis of Vision-based Abnormal Red Blood Cell Classification
Jun 01, 2021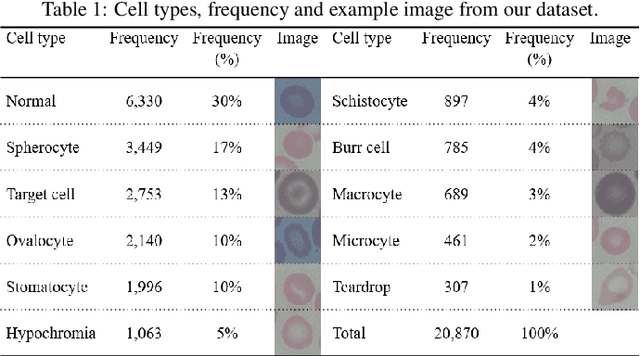
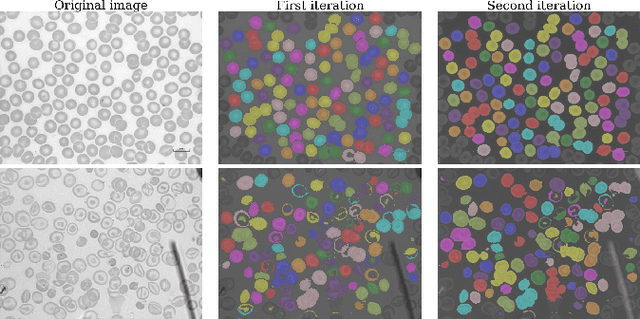
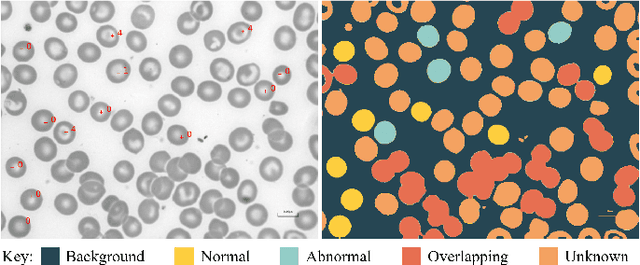
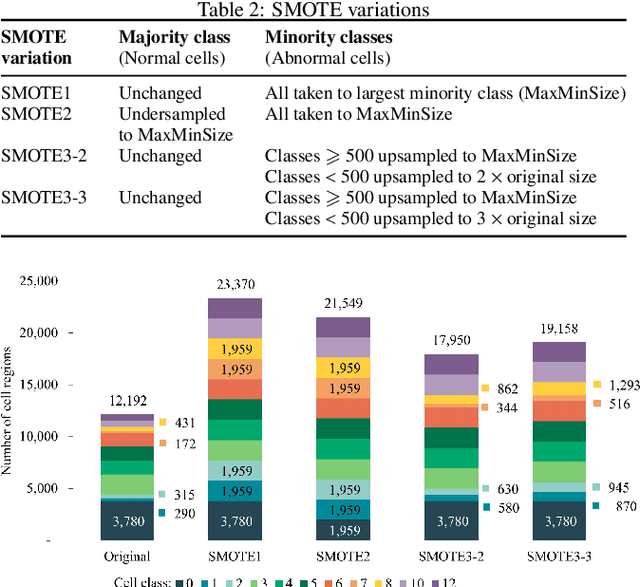
Abstract:Identification of abnormalities in red blood cells (RBC) is key to diagnosing a range of medical conditions from anaemia to liver disease. Currently this is done manually, a time-consuming and subjective process. This paper presents an automated process utilising the advantages of machine learning to increase capacity and standardisation of cell abnormality detection, and its performance is analysed. Three different machine learning technologies were used: a Support Vector Machine (SVM), a classical machine learning technology; TabNet, a deep learning architecture for tabular data; U-Net, a semantic segmentation network designed for medical image segmentation. A critical issue was the highly imbalanced nature of the dataset which impacts the efficacy of machine learning. To address this, synthesising minority class samples in feature space was investigated via Synthetic Minority Over-sampling Technique (SMOTE) and cost-sensitive learning. A combination of these two methods is investigated to improve the overall performance. These strategies were found to increase sensitivity to minority classes. The impact of unknown cells on semantic segmentation is demonstrated, with some evidence of the model applying learning of labelled cells to these anonymous cells. These findings indicate both classical models and new deep learning networks as promising methods in automating RBC abnormality detection.
Red Blood Cell Segmentation with Overlapping Cell Separation and Classification on Imbalanced Dataset
Dec 09, 2020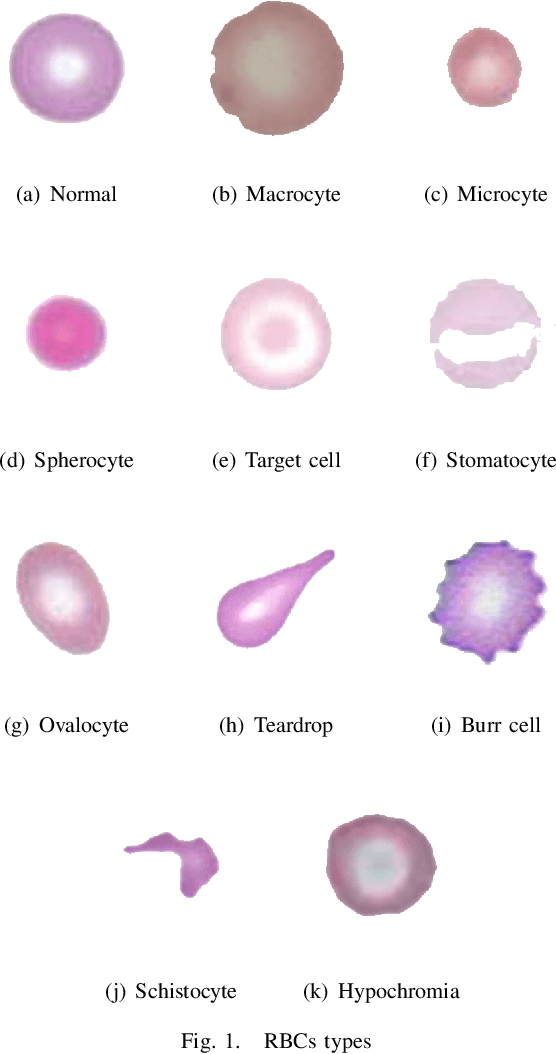
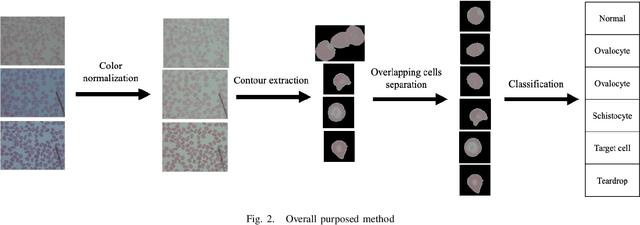
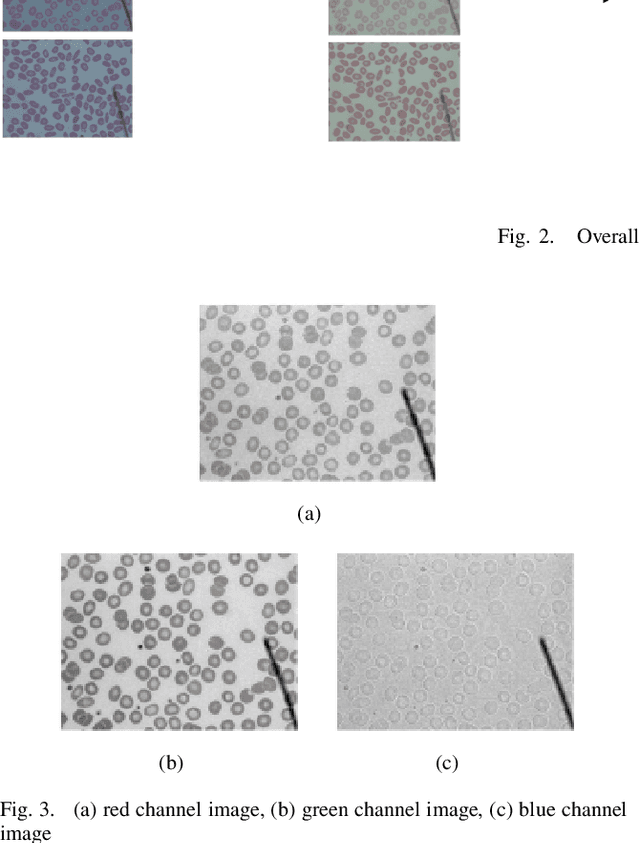
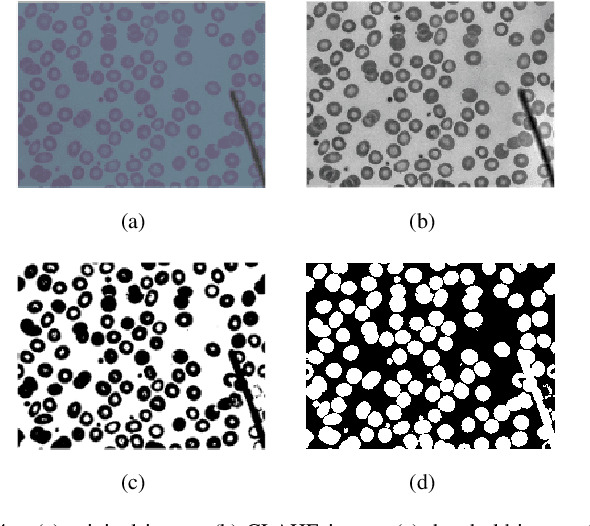
Abstract:Automated red blood cell classification on blood smear images helps hematologist to analyze RBC lab results in less time and cost. Overlapping cells can cause incorrect predicted results that have to separate into multiple single RBCs before classifying. To classify multiple classes with deep learning, imbalance problems are common in medical imaging because normal samples are always higher than rare disease samples. This paper presents a new method to segment and classify red blood cells from blood smear images, specifically to tackle cell overlapping and data imbalance problems. Focusing on overlapping cell separation, our segmentation process first estimates ellipses to represent red blood cells. The method detects the concave points and then finds the ellipses using directed ellipse fitting. The accuracy is 0.889 on 20 blood smear images. Classification requires balanced training datasets. However, some RBC types are rare. The imbalance ratio is 34.538 on 12 classes with 20,875 individual red blood cell samples. The use of machine learning for RBC classification with an imbalance dataset is hence more challenging than many other applications. We analyze techniques to deal with this problem. The best accuracy and f1 score are 0.921 and 0.8679 on EfficientNet-b1 with augmentation. Experimental results show that the weight balancing technique with augmentation has the potential to deal with imbalance problems by improving the f1 score on minority classes while data augmentation significantly improves the overall classification performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge