Annika Wong
Beyond Line-of-Sight: Cooperative Localization Using Vision and V2X Communication
Jul 28, 2025Abstract:Accurate and robust localization is critical for the safe operation of Connected and Automated Vehicles (CAVs), especially in complex urban environments where Global Navigation Satellite System (GNSS) signals are unreliable. This paper presents a novel vision-based cooperative localization algorithm that leverages onboard cameras and Vehicle-to-Everything (V2X) communication to enable CAVs to estimate their poses, even in occlusion-heavy scenarios such as busy intersections. In particular, we propose a novel decentralized observer for a group of connected agents that includes landmark agents (static or moving) in the environment with known positions and vehicle agents that need to estimate their poses (both positions and orientations). Assuming that (i) there are at least three landmark agents in the environment, (ii) each vehicle agent can measure its own angular and translational velocities as well as relative bearings to at least three neighboring landmarks or vehicles, and (iii) neighboring vehicles can communicate their pose estimates, each vehicle can estimate its own pose using the proposed decentralized observer. We prove that the origin of the estimation error is locally exponentially stable under the proposed observer, provided that the minimal observability conditions are satisfied. Moreover, we evaluate the proposed approach through experiments with real 1/10th-scale connected vehicles and large-scale simulations, demonstrating its scalability and validating the theoretical guarantees in practical scenarios.
Analysis of Vision-based Abnormal Red Blood Cell Classification
Jun 01, 2021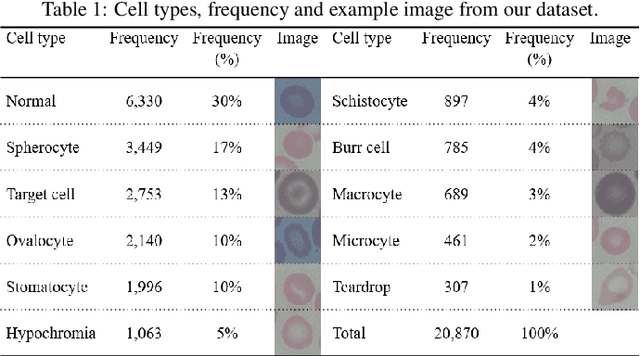
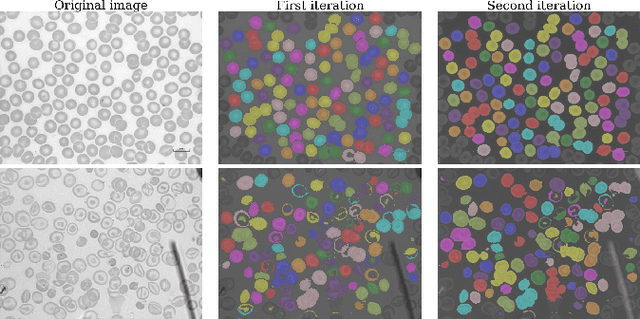
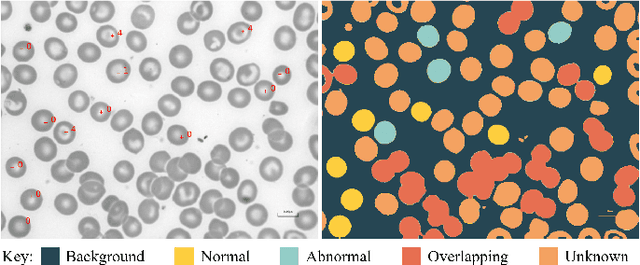
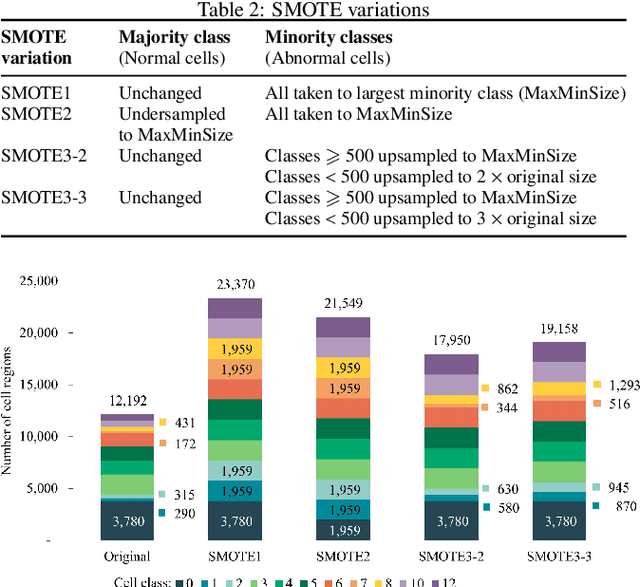
Abstract:Identification of abnormalities in red blood cells (RBC) is key to diagnosing a range of medical conditions from anaemia to liver disease. Currently this is done manually, a time-consuming and subjective process. This paper presents an automated process utilising the advantages of machine learning to increase capacity and standardisation of cell abnormality detection, and its performance is analysed. Three different machine learning technologies were used: a Support Vector Machine (SVM), a classical machine learning technology; TabNet, a deep learning architecture for tabular data; U-Net, a semantic segmentation network designed for medical image segmentation. A critical issue was the highly imbalanced nature of the dataset which impacts the efficacy of machine learning. To address this, synthesising minority class samples in feature space was investigated via Synthetic Minority Over-sampling Technique (SMOTE) and cost-sensitive learning. A combination of these two methods is investigated to improve the overall performance. These strategies were found to increase sensitivity to minority classes. The impact of unknown cells on semantic segmentation is demonstrated, with some evidence of the model applying learning of labelled cells to these anonymous cells. These findings indicate both classical models and new deep learning networks as promising methods in automating RBC abnormality detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge