Diana Waldmannstetter
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
Framing image registration as a landmark detection problem for better representation of clinical relevance
Jul 31, 2023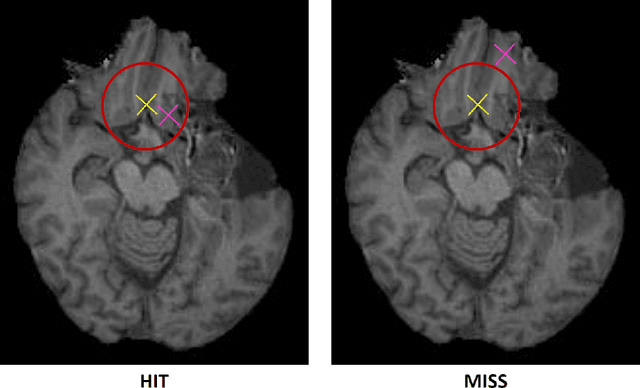
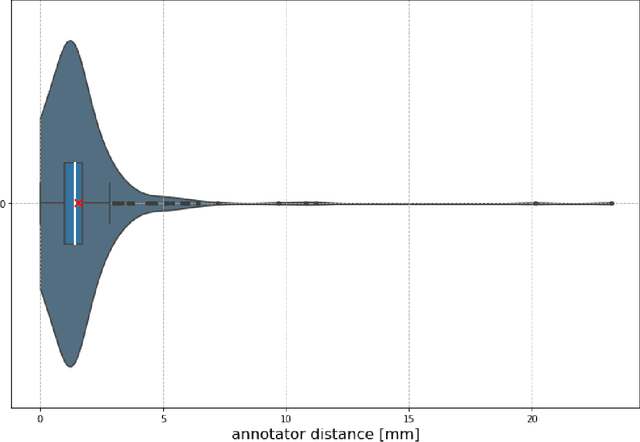
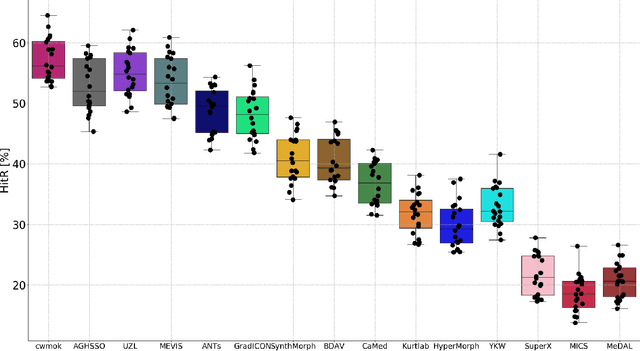
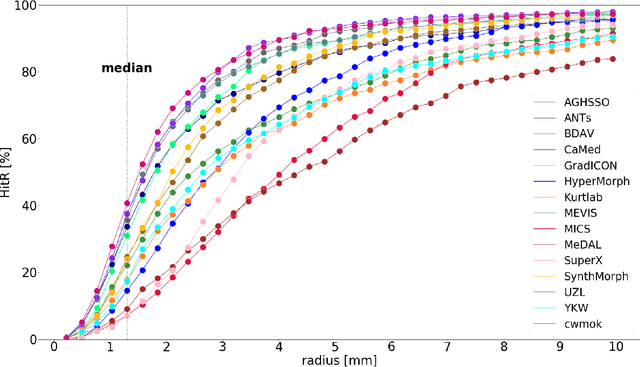
Abstract:Nowadays, registration methods are typically evaluated based on sub-resolution tracking error differences. In an effort to reinfuse this evaluation process with clinical relevance, we propose to reframe image registration as a landmark detection problem. Ideally, landmark-specific detection thresholds are derived from an inter-rater analysis. To approximate this costly process, we propose to compute hit rate curves based on the distribution of errors of a sub-sample inter-rater analysis. Therefore, we suggest deriving thresholds from the error distribution using the formula: median + delta * median absolute deviation. The method promises differentiation of previously indistinguishable registration algorithms and further enables assessing the clinical significance in algorithm development.
The Brain Tumor Segmentation Challenge 2023: Local Synthesis of Healthy Brain Tissue via Inpainting
May 15, 2023



Abstract:A myriad of algorithms for the automatic analysis of brain MR images is available to support clinicians in their decision-making. For brain tumor patients, the image acquisition time series typically starts with a scan that is already pathological. This poses problems, as many algorithms are designed to analyze healthy brains and provide no guarantees for images featuring lesions. Examples include but are not limited to algorithms for brain anatomy parcellation, tissue segmentation, and brain extraction. To solve this dilemma, we introduce the BraTS 2023 inpainting challenge. Here, the participants' task is to explore inpainting techniques to synthesize healthy brain scans from lesioned ones. The following manuscript contains the task formulation, dataset, and submission procedure. Later it will be updated to summarize the findings of the challenge. The challenge is organized as part of the BraTS 2023 challenge hosted at the MICCAI 2023 conference in Vancouver, Canada.
Primitive Simultaneous Optimization of Similarity Metrics for Image Registration
Apr 04, 2023



Abstract:Even though simultaneous optimization of similarity metrics represents a standard procedure in the field of semantic segmentation, surprisingly, this does not hold true for image registration. To close this unexpected gap in the literature, we investigate in a complex multi-modal 3D setting whether simultaneous optimization of registration metrics, here implemented by means of primitive summation, can benefit image registration. We evaluate two challenging datasets containing collections of pre- to post-operative and pre- to intra-operative Magnetic Resonance Imaging (MRI) of glioma. Employing the proposed optimization we demonstrate improved registration accuracy in terms of Target Registration Error (TRE) on expert neuroradiologists' landmark annotations.
The Brain Tumor Sequence Registration Challenge: Establishing Correspondence between Pre-Operative and Follow-up MRI scans of diffuse glioma patients
Dec 13, 2021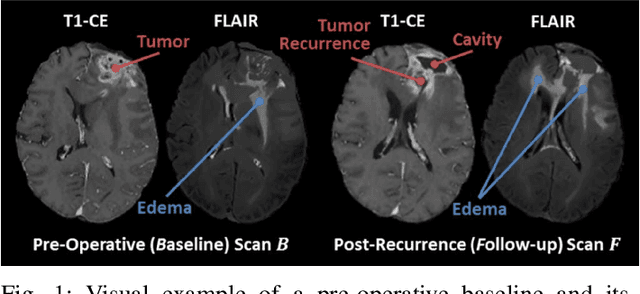
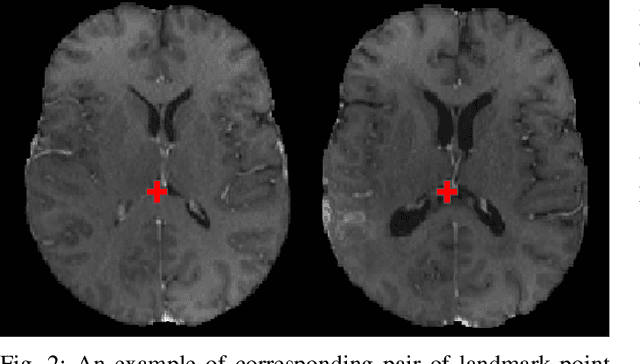
Abstract:Registration of longitudinal brain Magnetic Resonance Imaging (MRI) scans containing pathologies is challenging due to tissue appearance changes, and still an unsolved problem. This paper describes the first Brain Tumor Sequence Registration (BraTS-Reg) challenge, focusing on estimating correspondences between pre-operative and follow-up scans of the same patient diagnosed with a brain diffuse glioma. The BraTS-Reg challenge intends to establish a public benchmark environment for deformable registration algorithms. The associated dataset comprises de-identified multi-institutional multi-parametric MRI (mpMRI) data, curated for each scan's size and resolution, according to a common anatomical template. Clinical experts have generated extensive annotations of landmarks points within the scans, descriptive of distinct anatomical locations across the temporal domain. The training data along with these ground truth annotations will be released to participants to design and develop their registration algorithms, whereas the annotations for the validation and the testing data will be withheld by the organizers and used to evaluate the containerized algorithms of the participants. Each submitted algorithm will be quantitatively evaluated using several metrics, such as the Median Absolute Error (MAE), Robustness, and the Jacobian determinant.
Learn-Morph-Infer: a new way of solving the inverse problem for brain tumor modeling
Nov 07, 2021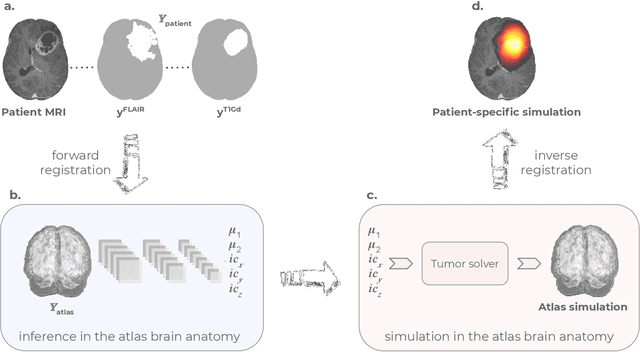
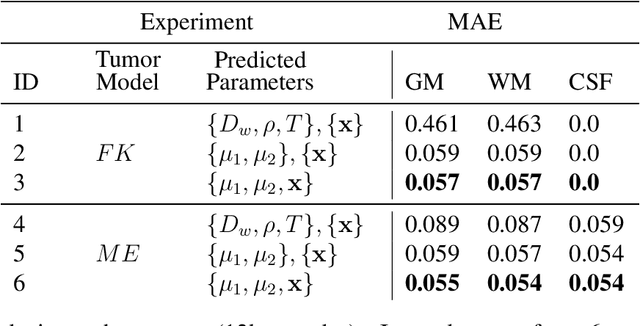
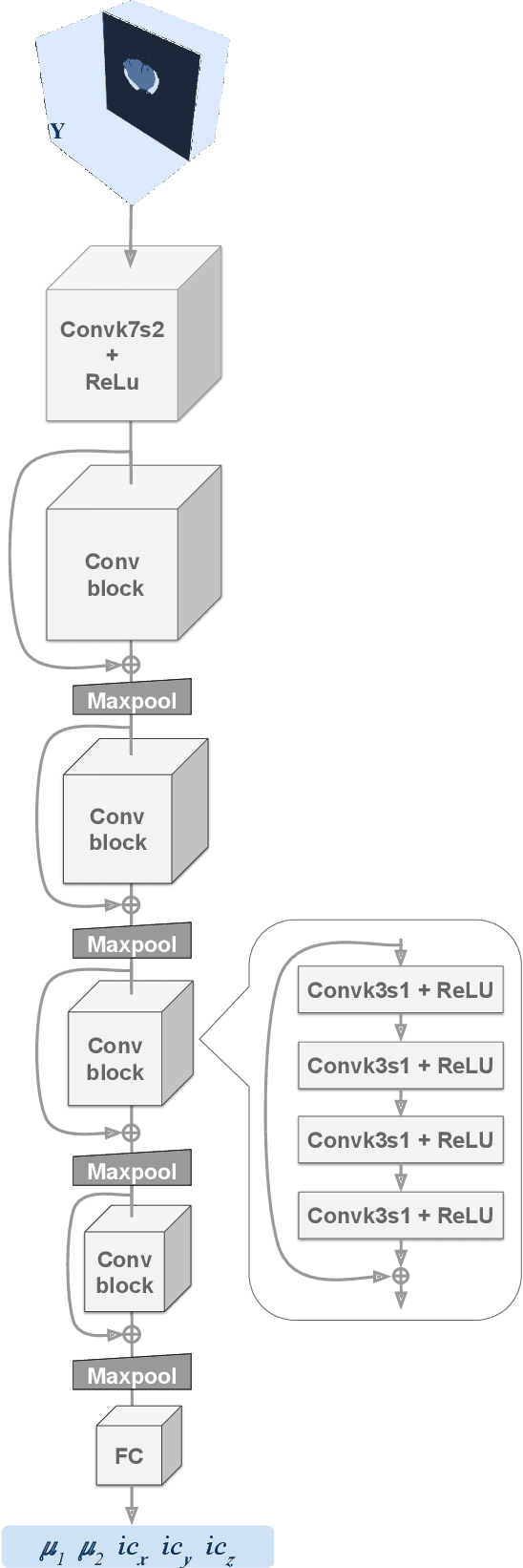
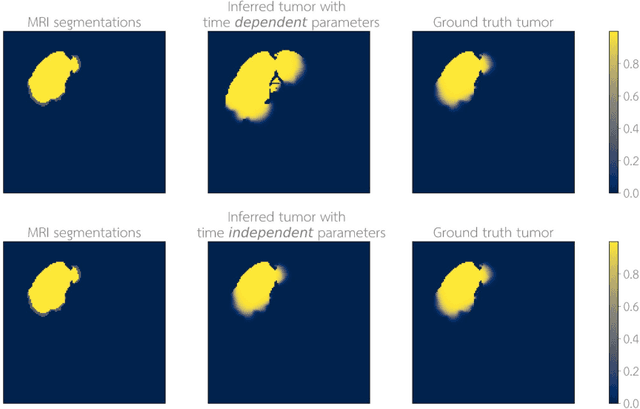
Abstract:Current treatment planning of patients diagnosed with brain tumor could significantly benefit by accessing the spatial distribution of tumor cell concentration. Existing diagnostic modalities, such as magnetic-resonance imaging (MRI), contrast sufficiently well areas of high cell density. However, they do not portray areas of low concentration, which can often serve as a source for the secondary appearance of the tumor after treatment. Numerical simulations of tumor growth could complement imaging information by providing estimates of full spatial distributions of tumor cells. Over recent years a corpus of literature on medical image-based tumor modeling was published. It includes different mathematical formalisms describing the forward tumor growth model. Alongside, various parametric inference schemes were developed to perform an efficient tumor model personalization, i.e. solving the inverse problem. However, the unifying drawback of all existing approaches is the time complexity of the model personalization that prohibits a potential integration of the modeling into clinical settings. In this work, we introduce a methodology for inferring patient-specific spatial distribution of brain tumor from T1Gd and FLAIR MRI medical scans. Coined as \textit{Learn-Morph-Infer} the method achieves real-time performance in the order of minutes on widely available hardware and the compute time is stable across tumor models of different complexity, such as reaction-diffusion and reaction-advection-diffusion models. We believe the proposed inverse solution approach not only bridges the way for clinical translation of brain tumor personalization but can also be adopted to other scientific and engineering domains.
Deep Reinforcement Learning for Organ Localization in CT
May 11, 2020
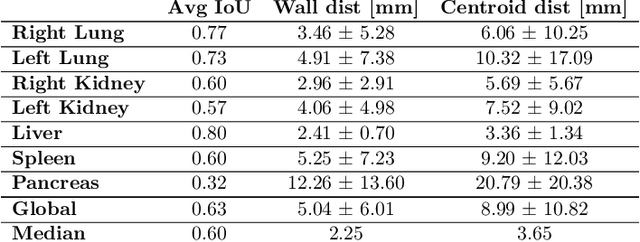

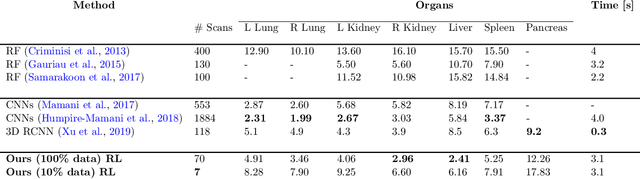
Abstract:Robust localization of organs in computed tomography scans is a constant pre-processing requirement for organ-specific image retrieval, radiotherapy planning, and interventional image analysis. In contrast to current solutions based on exhaustive search or region proposals, which require large amounts of annotated data, we propose a deep reinforcement learning approach for organ localization in CT. In this work, an artificial agent is actively self-taught to localize organs in CT by learning from its asserts and mistakes. Within the context of reinforcement learning, we propose a novel set of actions tailored for organ localization in CT. Our method can use as a plug-and-play module for localizing any organ of interest. We evaluate the proposed solution on the public VISCERAL dataset containing CT scans with varying fields of view and multiple organs. We achieved an overall intersection over union of 0.63, an absolute median wall distance of 2.25 mm, and a median distance between centroids of 3.65 mm.
* Accepted paper in MIDL 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge