Marie Metz
Framing image registration as a landmark detection problem for better representation of clinical relevance
Jul 31, 2023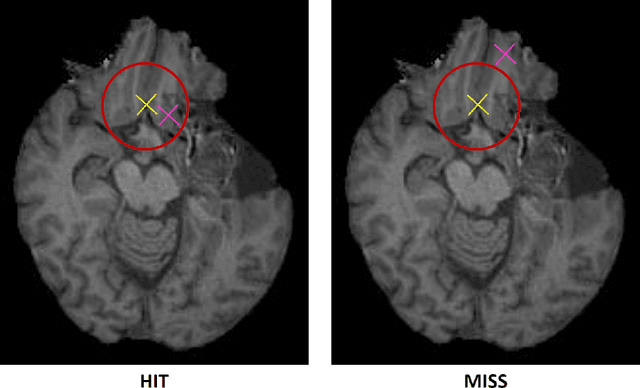
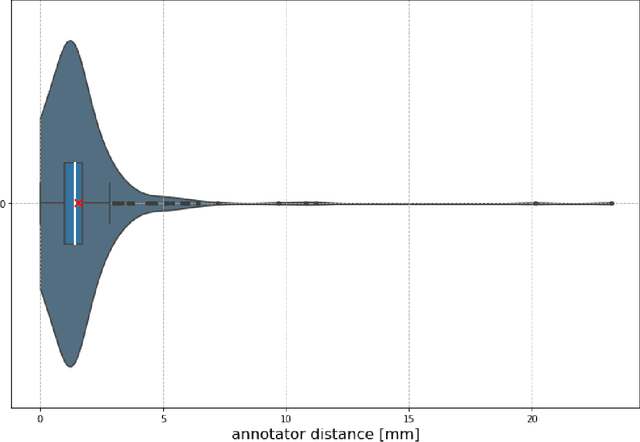
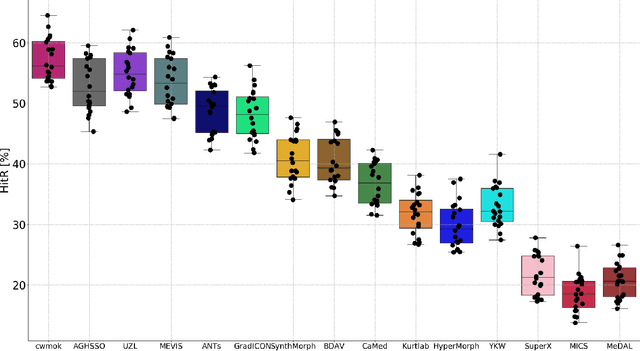
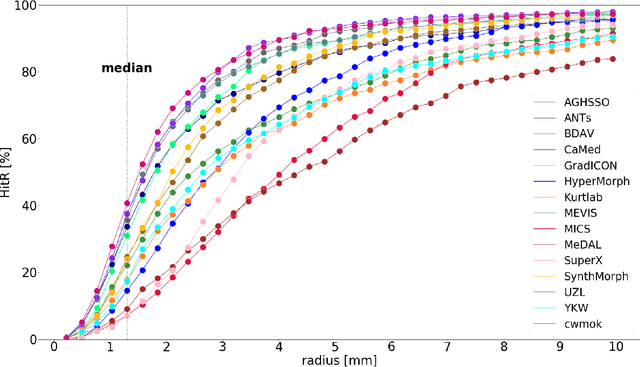
Abstract:Nowadays, registration methods are typically evaluated based on sub-resolution tracking error differences. In an effort to reinfuse this evaluation process with clinical relevance, we propose to reframe image registration as a landmark detection problem. Ideally, landmark-specific detection thresholds are derived from an inter-rater analysis. To approximate this costly process, we propose to compute hit rate curves based on the distribution of errors of a sub-sample inter-rater analysis. Therefore, we suggest deriving thresholds from the error distribution using the formula: median + delta * median absolute deviation. The method promises differentiation of previously indistinguishable registration algorithms and further enables assessing the clinical significance in algorithm development.
Primitive Simultaneous Optimization of Similarity Metrics for Image Registration
Apr 04, 2023



Abstract:Even though simultaneous optimization of similarity metrics represents a standard procedure in the field of semantic segmentation, surprisingly, this does not hold true for image registration. To close this unexpected gap in the literature, we investigate in a complex multi-modal 3D setting whether simultaneous optimization of registration metrics, here implemented by means of primitive summation, can benefit image registration. We evaluate two challenging datasets containing collections of pre- to post-operative and pre- to intra-operative Magnetic Resonance Imaging (MRI) of glioma. Employing the proposed optimization we demonstrate improved registration accuracy in terms of Target Registration Error (TRE) on expert neuroradiologists' landmark annotations.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Learn-Morph-Infer: a new way of solving the inverse problem for brain tumor modeling
Nov 07, 2021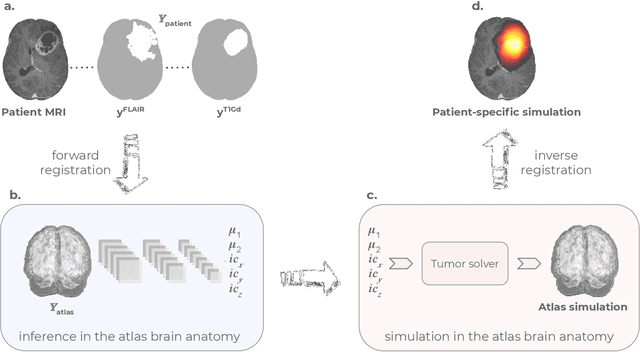
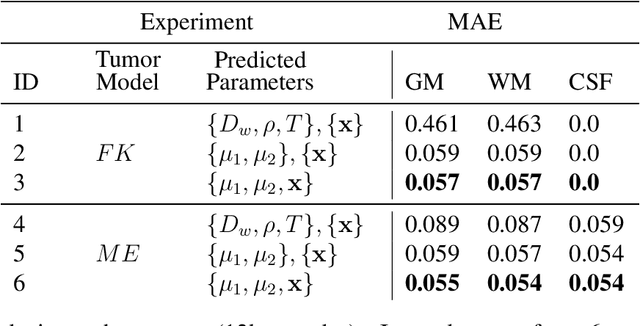
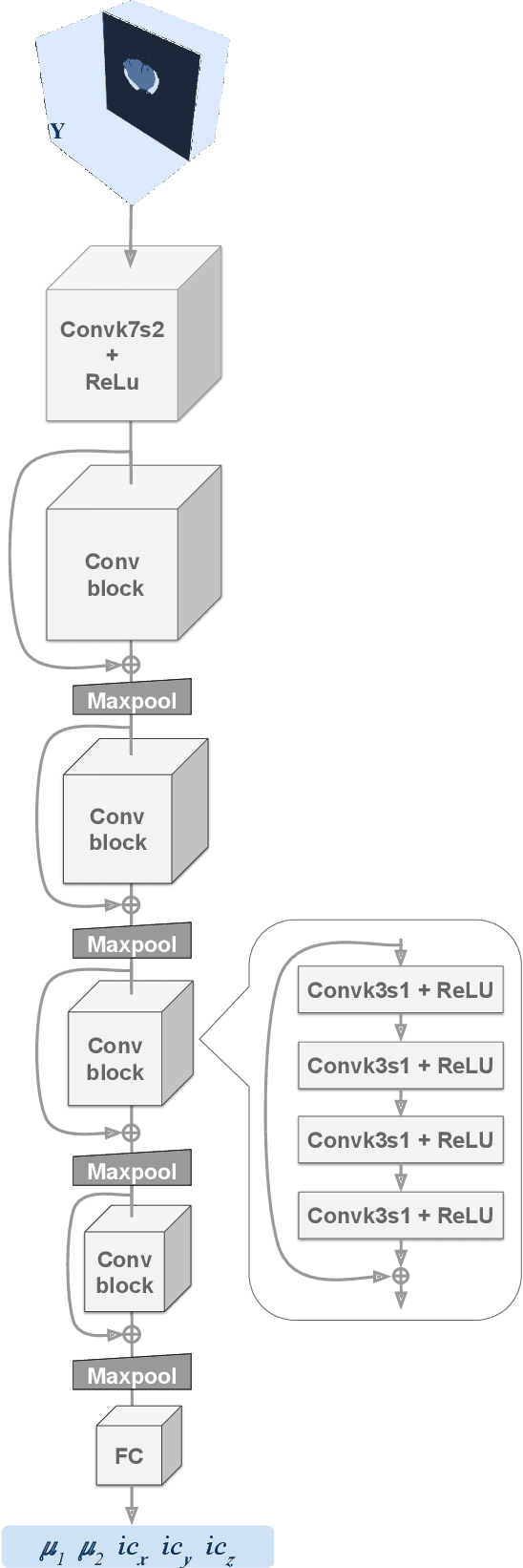
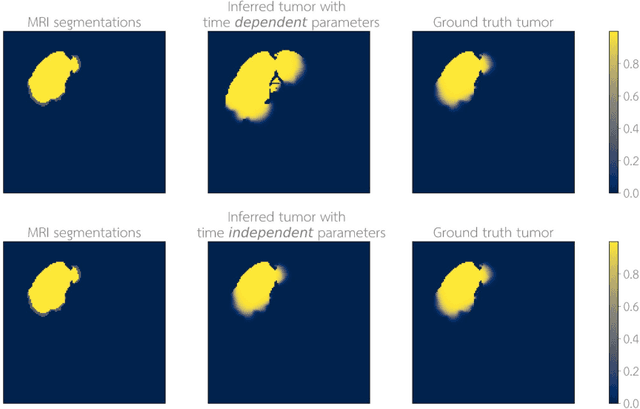
Abstract:Current treatment planning of patients diagnosed with brain tumor could significantly benefit by accessing the spatial distribution of tumor cell concentration. Existing diagnostic modalities, such as magnetic-resonance imaging (MRI), contrast sufficiently well areas of high cell density. However, they do not portray areas of low concentration, which can often serve as a source for the secondary appearance of the tumor after treatment. Numerical simulations of tumor growth could complement imaging information by providing estimates of full spatial distributions of tumor cells. Over recent years a corpus of literature on medical image-based tumor modeling was published. It includes different mathematical formalisms describing the forward tumor growth model. Alongside, various parametric inference schemes were developed to perform an efficient tumor model personalization, i.e. solving the inverse problem. However, the unifying drawback of all existing approaches is the time complexity of the model personalization that prohibits a potential integration of the modeling into clinical settings. In this work, we introduce a methodology for inferring patient-specific spatial distribution of brain tumor from T1Gd and FLAIR MRI medical scans. Coined as \textit{Learn-Morph-Infer} the method achieves real-time performance in the order of minutes on widely available hardware and the compute time is stable across tumor models of different complexity, such as reaction-diffusion and reaction-advection-diffusion models. We believe the proposed inverse solution approach not only bridges the way for clinical translation of brain tumor personalization but can also be adopted to other scientific and engineering domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge