Hamed Akbari
Artificial intelligence-based locoregional markers of brain peritumoral microenvironment
Aug 29, 2022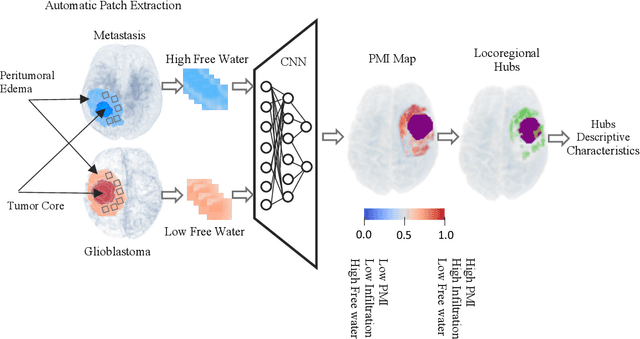
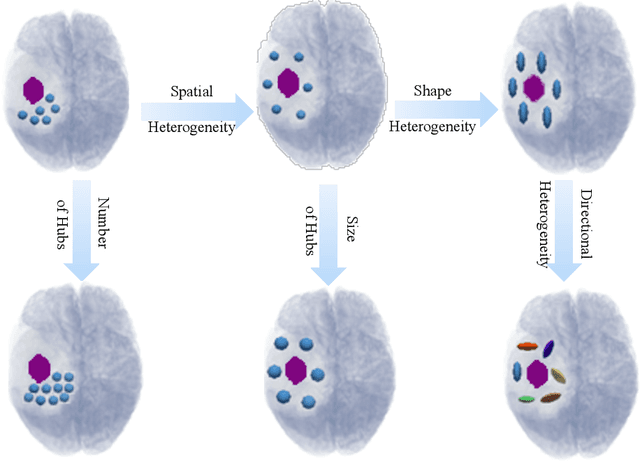
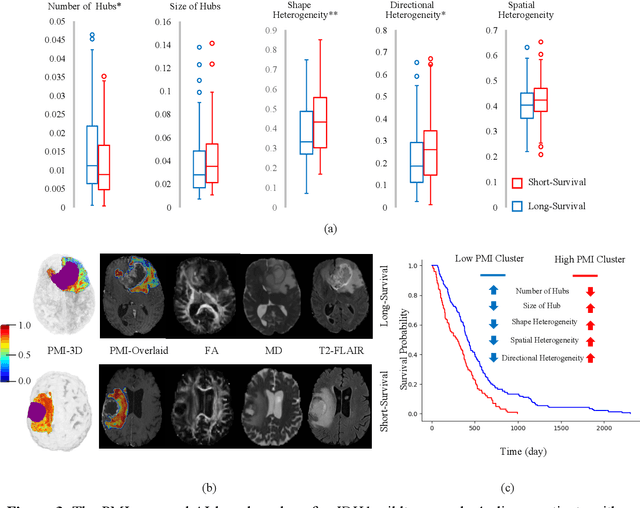
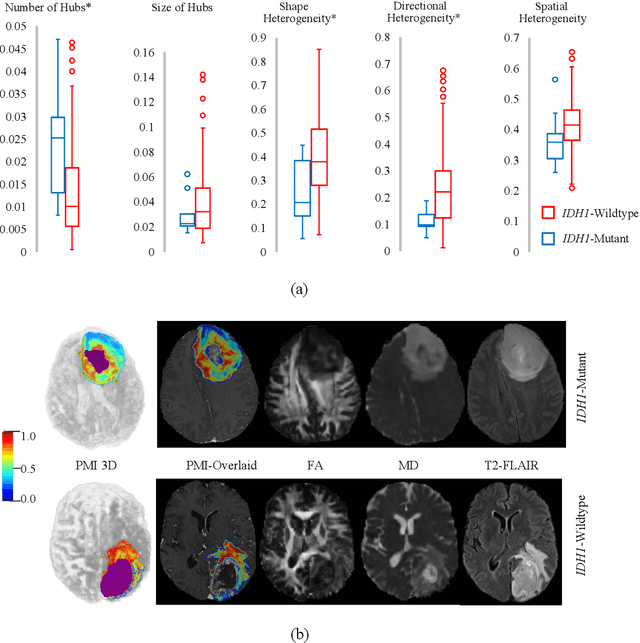
Abstract:In malignant primary brain tumors, cancer cells infiltrate into the peritumoral brain structures which results in inevitable recurrence. Quantitative assessment of infiltrative heterogeneity in the peritumoral region, the area where biopsy or resection can be hazardous, is important for clinical decision making. Previous work on characterizing the infiltrative heterogeneity in the peritumoral region used various imaging modalities, but information of extracellular free water movement restriction has been limitedly explored. Here, we derive a unique set of Artificial Intelligence (AI)-based markers capturing the heterogeneity of tumor infiltration, by characterizing free water movement restriction in the peritumoral region using Diffusion Tensor Imaging (DTI)-based free water volume fraction maps. A novel voxel-wise deep learning-based peritumoral microenvironment index (PMI) is first extracted by leveraging the widely different water diffusivity properties of glioblastomas and brain metastases as regions with and without infiltrations in the peritumoral tissue. Descriptive characteristics of locoregional hubs of uniformly high PMI values are extracted as AI-based markers to capture distinct aspects of infiltrative heterogeneity. The proposed markers are applied to two clinical use cases on an independent population of 275 adult-type diffuse gliomas (CNS WHO grade 4), analyzing the duration of survival among Isocitrate-Dehydrogenase 1 (IDH1)-wildtypes and the differences with IDH1-mutants. Our findings provide a panel of markers as surrogates of infiltration that captures unique insight about underlying biology of peritumoral microstructural heterogeneity, establishing them as biomarkers of prognosis pertaining to survival and molecular stratification, with potential applicability in clinical decision making.
The Brain Tumor Sequence Registration Challenge: Establishing Correspondence between Pre-Operative and Follow-up MRI scans of diffuse glioma patients
Dec 13, 2021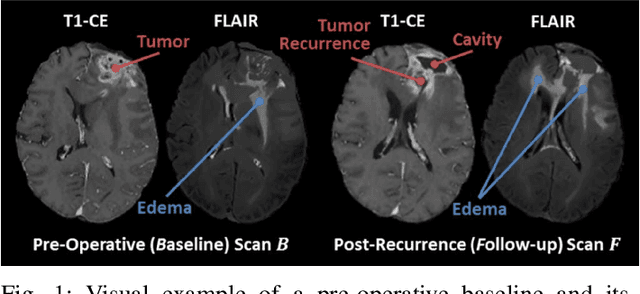
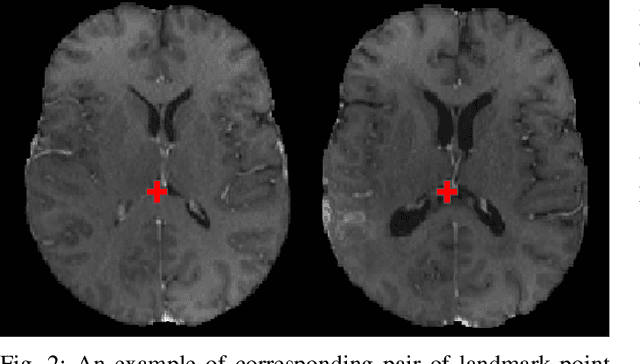
Abstract:Registration of longitudinal brain Magnetic Resonance Imaging (MRI) scans containing pathologies is challenging due to tissue appearance changes, and still an unsolved problem. This paper describes the first Brain Tumor Sequence Registration (BraTS-Reg) challenge, focusing on estimating correspondences between pre-operative and follow-up scans of the same patient diagnosed with a brain diffuse glioma. The BraTS-Reg challenge intends to establish a public benchmark environment for deformable registration algorithms. The associated dataset comprises de-identified multi-institutional multi-parametric MRI (mpMRI) data, curated for each scan's size and resolution, according to a common anatomical template. Clinical experts have generated extensive annotations of landmarks points within the scans, descriptive of distinct anatomical locations across the temporal domain. The training data along with these ground truth annotations will be released to participants to design and develop their registration algorithms, whereas the annotations for the validation and the testing data will be withheld by the organizers and used to evaluate the containerized algorithms of the participants. Each submitted algorithm will be quantitatively evaluated using several metrics, such as the Median Absolute Error (MAE), Robustness, and the Jacobian determinant.
A Deep Network for Joint Registration and Reconstruction of Images with Pathologies
Aug 17, 2020



Abstract:Registration of images with pathologies is challenging due to tissue appearance changes and missing correspondences caused by the pathologies. Moreover, mass effects as observed for brain tumors may displace tissue, creating larger deformations over time than what is observed in a healthy brain. Deep learning models have successfully been applied to image registration to offer dramatic speed up and to use surrogate information (e.g., segmentations) during training. However, existing approaches focus on learning registration models using images from healthy patients. They are therefore not designed for the registration of images with strong pathologies for example in the context of brain tumors, and traumatic brain injuries. In this work, we explore a deep learning approach to register images with brain tumors to an atlas. Our model learns an appearance mapping from images with tumors to the atlas, while simultaneously predicting the transformation to atlas space. Using separate decoders, the network disentangles the tumor mass effect from the reconstruction of quasi-normal images. Results on both synthetic and real brain tumor scans show that our approach outperforms cost function masking for registration to the atlas and that reconstructed quasi-normal images can be used for better longitudinal registrations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge