Ragini Verma
Parsing altered brain connectivity in neurodevelopmental disorders by integrating graph-based normative modeling and deep generative networks
Oct 14, 2024Abstract:Many neurodevelopmental disorders can be understood as divergent patterns of neural interactions during brain development. Advances in neuroimaging have illuminated these patterns by modeling the brain as a network structure using diffution MRI tractography. However, characterizing and quantifying individual heterogeneity in neurodevelopmental disorders within these highly complex brain networks remains a significant challenge. In this paper, we present for the first time, a framework that integrates deep generative models with graph-based normative modeling to characterize brain network development in the neurotypical population, which can then be used to quantify the individual-level neurodivergence associated with disorders. Our deep generative model incorporates bio-inspired wiring constraints to effectively capture the developmental trajectories of neurotypical brain networks. Neurodivergence is quantified by comparing individuals to this neurotypical trajectory, enabling the creation of region-wise divergence maps that reveal latent developmental differences at each brain regions, along with overall neurodivergence scores based on predicted brain age gaps. We demonstrate the clinical utility of this framework by applying it to a large sample of children with autism spectrum disorders, showing that the individualized region-wise maps help parse the heterogeneity in autism, and the neurodivergence scores correlate with clinical assessments. Together, we provide powerful tools for quantifying neurodevelopmental divergence in brain networks, paying the way for developing imaging markers that will support disorder stratification, monitor progression, and evaluate therapeutic effectiveness.
Linking Symptom Inventories using Semantic Textual Similarity
Sep 08, 2023
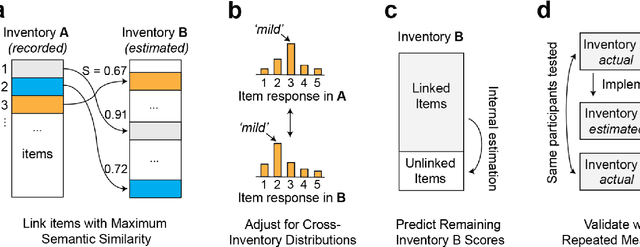
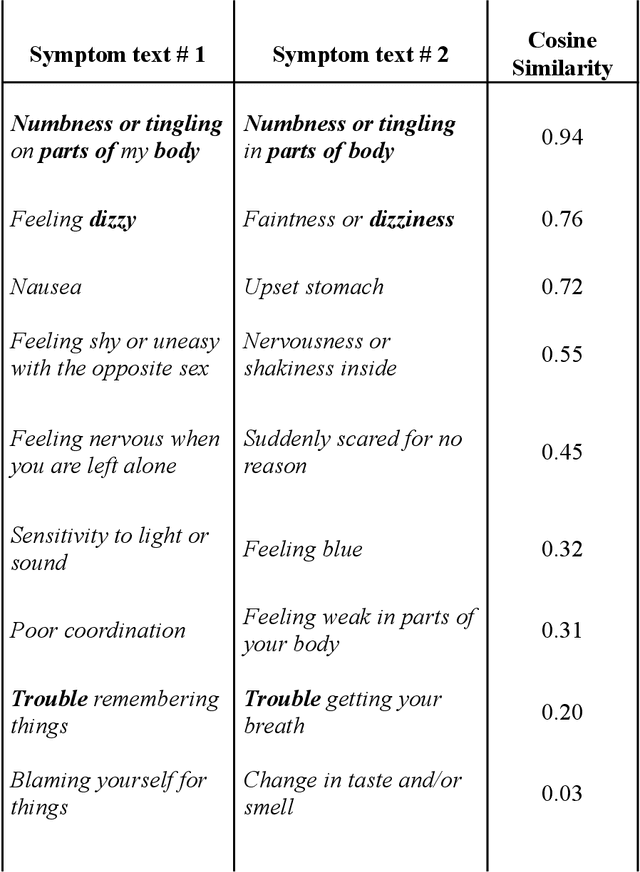
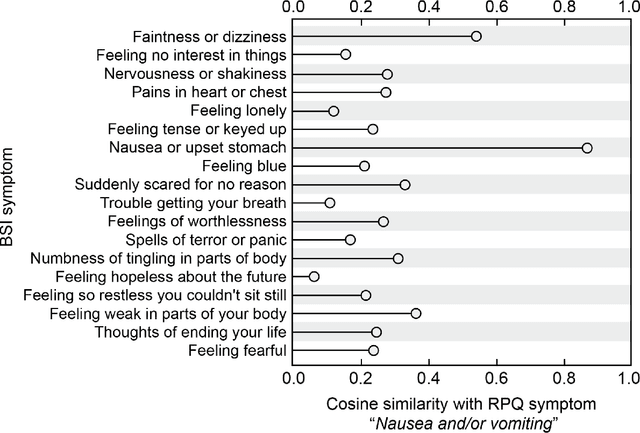
Abstract:An extensive library of symptom inventories has been developed over time to measure clinical symptoms, but this variety has led to several long standing issues. Most notably, results drawn from different settings and studies are not comparable, which limits reproducibility. Here, we present an artificial intelligence (AI) approach using semantic textual similarity (STS) to link symptoms and scores across previously incongruous symptom inventories. We tested the ability of four pre-trained STS models to screen thousands of symptom description pairs for related content - a challenging task typically requiring expert panels. Models were tasked to predict symptom severity across four different inventories for 6,607 participants drawn from 16 international data sources. The STS approach achieved 74.8% accuracy across five tasks, outperforming other models tested. This work suggests that incorporating contextual, semantic information can assist expert decision-making processes, yielding gains for both general and disease-specific clinical assessment.
Artificial intelligence-based locoregional markers of brain peritumoral microenvironment
Aug 29, 2022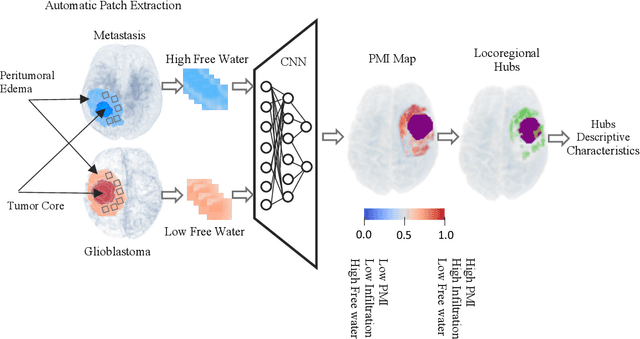
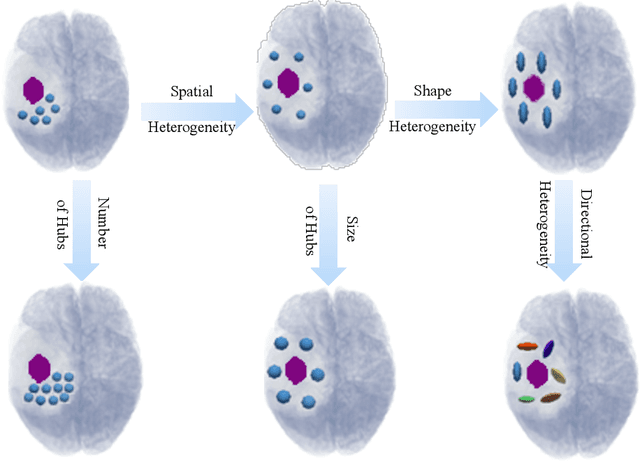
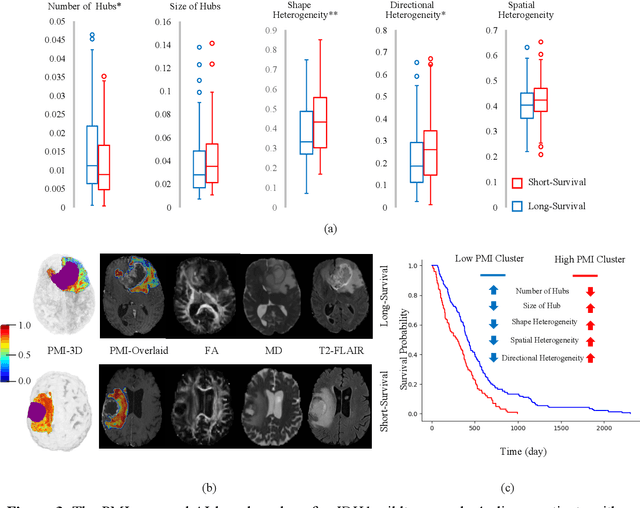
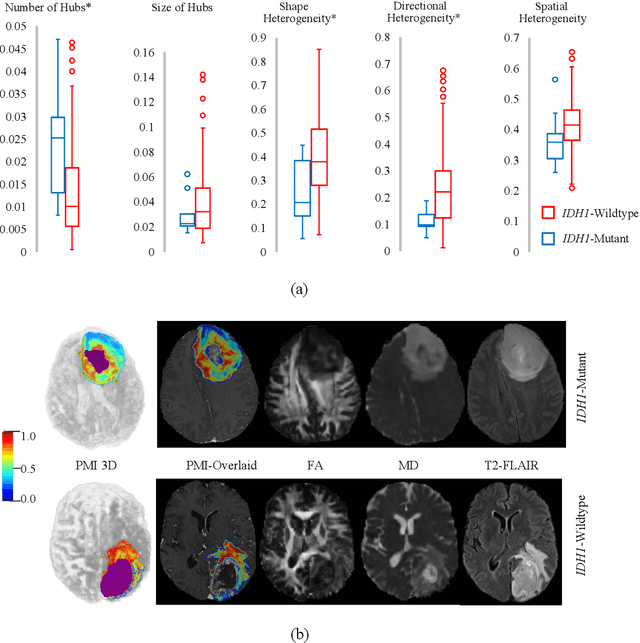
Abstract:In malignant primary brain tumors, cancer cells infiltrate into the peritumoral brain structures which results in inevitable recurrence. Quantitative assessment of infiltrative heterogeneity in the peritumoral region, the area where biopsy or resection can be hazardous, is important for clinical decision making. Previous work on characterizing the infiltrative heterogeneity in the peritumoral region used various imaging modalities, but information of extracellular free water movement restriction has been limitedly explored. Here, we derive a unique set of Artificial Intelligence (AI)-based markers capturing the heterogeneity of tumor infiltration, by characterizing free water movement restriction in the peritumoral region using Diffusion Tensor Imaging (DTI)-based free water volume fraction maps. A novel voxel-wise deep learning-based peritumoral microenvironment index (PMI) is first extracted by leveraging the widely different water diffusivity properties of glioblastomas and brain metastases as regions with and without infiltrations in the peritumoral tissue. Descriptive characteristics of locoregional hubs of uniformly high PMI values are extracted as AI-based markers to capture distinct aspects of infiltrative heterogeneity. The proposed markers are applied to two clinical use cases on an independent population of 275 adult-type diffuse gliomas (CNS WHO grade 4), analyzing the duration of survival among Isocitrate-Dehydrogenase 1 (IDH1)-wildtypes and the differences with IDH1-mutants. Our findings provide a panel of markers as surrogates of infiltration that captures unique insight about underlying biology of peritumoral microstructural heterogeneity, establishing them as biomarkers of prognosis pertaining to survival and molecular stratification, with potential applicability in clinical decision making.
3D-QCNet -- A Pipeline for Automated Artifact Detection in Diffusion MRI images
Mar 09, 2021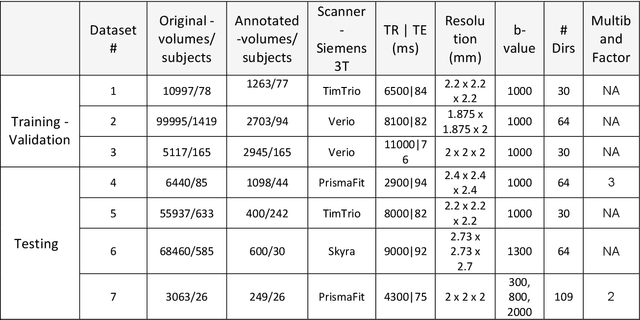
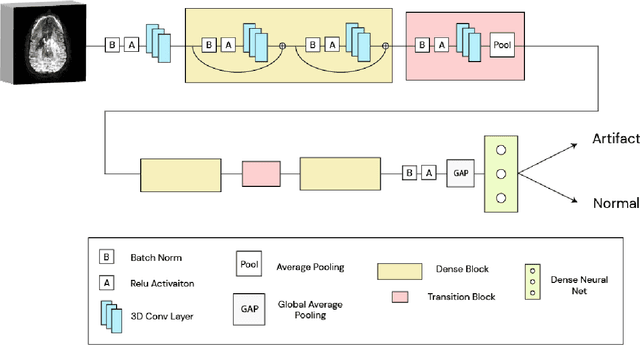
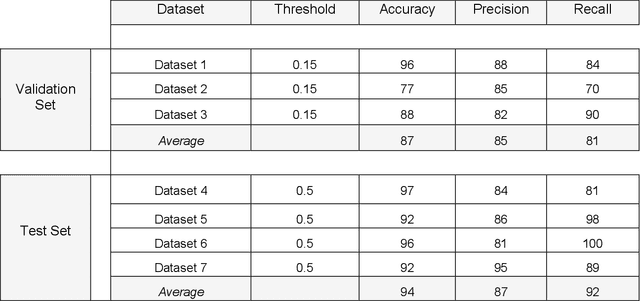
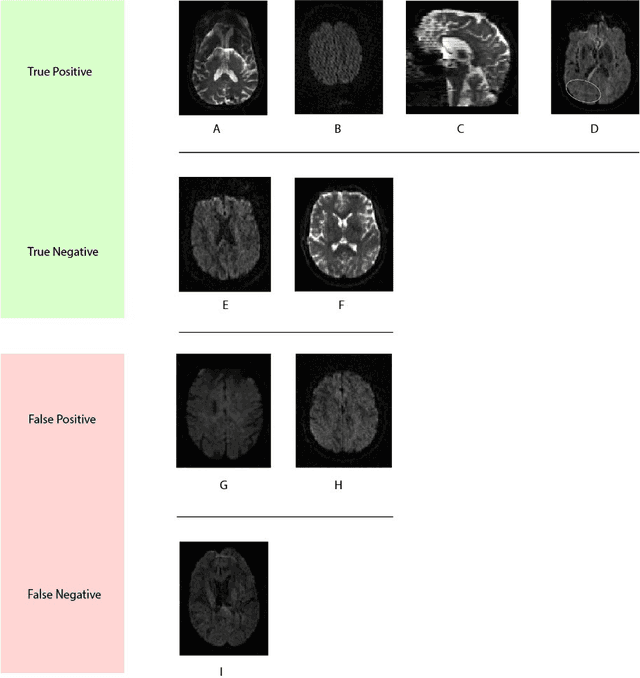
Abstract:Artifacts are a common occurrence in Diffusion MRI (dMRI) scans. Identifying and removing them is essential to ensure the accuracy and viability of any post processing carried out on these scans. This makes QC (quality control) a crucial first step prior to any analysis of dMRI data. Several QC methods for artifact detection exist, however they suffer from problems like requiring manual intervention and the inability to generalize across different artifacts and datasets. In this paper, we propose an automated deep learning (DL) pipeline that utilizes a 3D-Densenet architecture to train a model on diffusion volumes for automatic artifact detection. Our method is applied on a vast dataset consisting of 9000 volumes sourced from 7 large clinical datasets. These datasets comprise scans from multiple scanners with different gradient directions, high and low b values, single shell and multi shell acquisitions. Additionally, they represent diverse subject demographics like the presence or absence of pathologies. Our QC method is found to accurately generalize across this heterogenous data by correctly detecting 92% artifacts on average across our test set. This consistent performance over diverse datasets underlines the generalizability of our method, which currently is a significant barrier hindering the widespread adoption of automated QC techniques. For these reasons, we believe that 3D-QCNet can be integrated in diffusion pipelines to effectively automate the arduous and time-intensive process of artifact detection.
QC-Automator: Deep Learning-based Automated Quality Control for Diffusion MR Images
Nov 15, 2019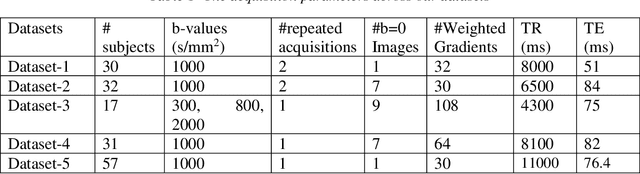
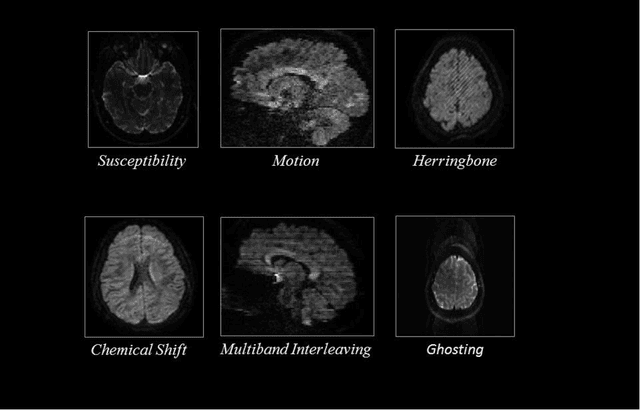
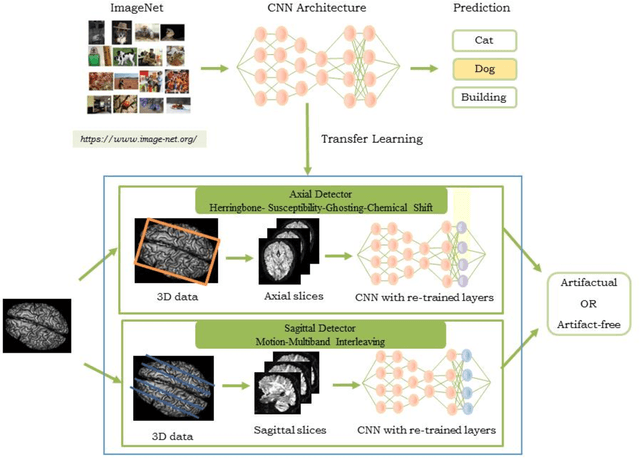
Abstract:Quality assessment of diffusion MRI (dMRI) data is essential prior to any analysis, so that appropriate pre-processing can be used to improve data quality and ensure that the presence of MRI artifacts do not affect the results of subsequent image analysis. Manual quality assessment of the data is subjective, possibly error-prone, and infeasible, especially considering the growing number of consortium-like studies, underlining the need for automation of the process. In this paper, we have developed a deep-learning-based automated quality control (QC) tool, QC-Automator, for dMRI data, that can handle a variety of artifacts such as motion, multiband interleaving, ghosting, susceptibility, herringbone and chemical shifts. QC-Automator uses convolutional neural networks along with transfer learning to train the automated artifact detection on a labeled dataset of ~332000 slices of dMRI data, from 155 unique subjects and 5 scanners with different dMRI acquisitions, achieving a 98% accuracy in detecting artifacts. The method is fast and paves the way for efficient and effective artifact detection in large datasets. It is also demonstrated to be replicable on other datasets with different acquisition parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge