Beatrice Alex
Adverse Event Extraction from Discharge Summaries: A New Dataset, Annotation Scheme, and Initial Findings
Jun 17, 2025Abstract:In this work, we present a manually annotated corpus for Adverse Event (AE) extraction from discharge summaries of elderly patients, a population often underrepresented in clinical NLP resources. The dataset includes 14 clinically significant AEs-such as falls, delirium, and intracranial haemorrhage, along with contextual attributes like negation, diagnosis type, and in-hospital occurrence. Uniquely, the annotation schema supports both discontinuous and overlapping entities, addressing challenges rarely tackled in prior work. We evaluate multiple models using FlairNLP across three annotation granularities: fine-grained, coarse-grained, and coarse-grained with negation. While transformer-based models (e.g., BERT-cased) achieve strong performance on document-level coarse-grained extraction (F1 = 0.943), performance drops notably for fine-grained entity-level tasks (e.g., F1 = 0.675), particularly for rare events and complex attributes. These results demonstrate that despite high-level scores, significant challenges remain in detecting underrepresented AEs and capturing nuanced clinical language. Developed within a Trusted Research Environment (TRE), the dataset is available upon request via DataLoch and serves as a robust benchmark for evaluating AE extraction methods and supporting future cross-dataset generalisation.
Investigating the Capabilities and Limitations of Machine Learning for Identifying Bias in English Language Data with Information and Heritage Professionals
Apr 01, 2025Abstract:Despite numerous efforts to mitigate their biases, ML systems continue to harm already-marginalized people. While predominant ML approaches assume bias can be removed and fair models can be created, we show that these are not always possible, nor desirable, goals. We reframe the problem of ML bias by creating models to identify biased language, drawing attention to a dataset's biases rather than trying to remove them. Then, through a workshop, we evaluated the models for a specific use case: workflows of information and heritage professionals. Our findings demonstrate the limitations of ML for identifying bias due to its contextual nature, the way in which approaches to mitigating it can simultaneously privilege and oppress different communities, and its inevitability. We demonstrate the need to expand ML approaches to bias and fairness, providing a mixed-methods approach to investigating the feasibility of removing bias or achieving fairness in a given ML use case.
Evaluating and Adapting Large Language Models to Represent Folktales in Low-Resource Languages
Nov 08, 2024



Abstract:Folktales are a rich resource of knowledge about the society and culture of a civilisation. Digital folklore research aims to use automated techniques to better understand these folktales, and it relies on abstract representations of the textual data. Although a number of large language models (LLMs) claim to be able to represent low-resource langauges such as Irish and Gaelic, we present two classification tasks to explore how useful these representations are, and three adaptations to improve the performance of these models. We find that adapting the models to work with longer sequences, and continuing pre-training on the domain of folktales improves classification performance, although these findings are tempered by the impressive performance of a baseline SVM with non-contextual features.
DeCoRe: Decoding by Contrasting Retrieval Heads to Mitigate Hallucinations
Oct 24, 2024



Abstract:Large Language Models (LLMs) often hallucinate, producing unfaithful or factually incorrect outputs by misrepresenting the provided context or incorrectly recalling internal knowledge. Recent studies have identified specific attention heads within the Transformer architecture, known as retrieval heads, responsible for extracting relevant contextual information. We hypothesise that masking these retrieval heads can induce hallucinations and that contrasting the outputs of the base LLM and the masked LLM can reduce hallucinations. To this end, we propose Decoding by Contrasting Retrieval Heads (DeCoRe), a novel training-free decoding strategy that amplifies information found in the context and model parameters. DeCoRe mitigates potentially hallucinated responses by dynamically contrasting the outputs of the base LLM and the masked LLM, using conditional entropy as a guide. Our extensive experiments confirm that DeCoRe significantly improves performance on tasks requiring high contextual faithfulness, such as summarisation (XSum by 18.6%), instruction following (MemoTrap by 10.9%), and open-book question answering (NQ-Open by 2.4% and NQ-Swap by 5.5%).
Automated neuroradiological support systems for multiple cerebrovascular disease markers -- A systematic review and meta-analysis
Oct 22, 2024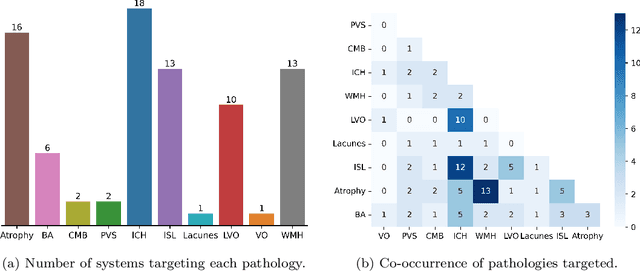
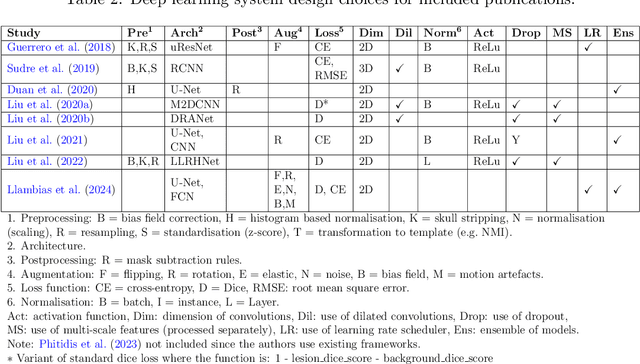
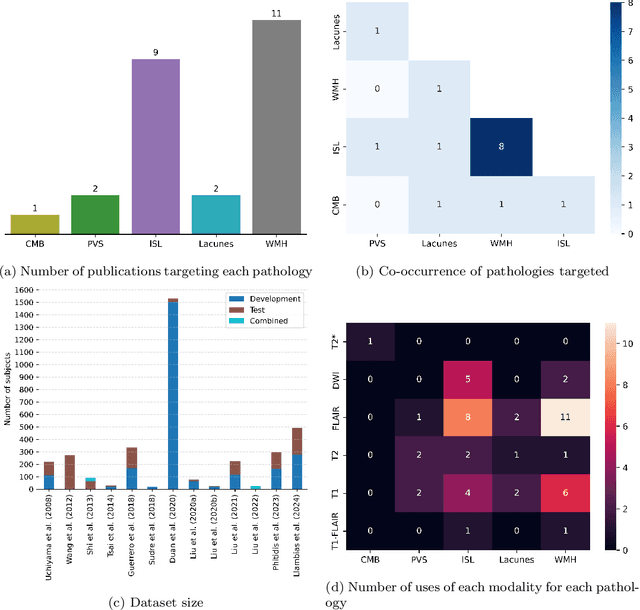
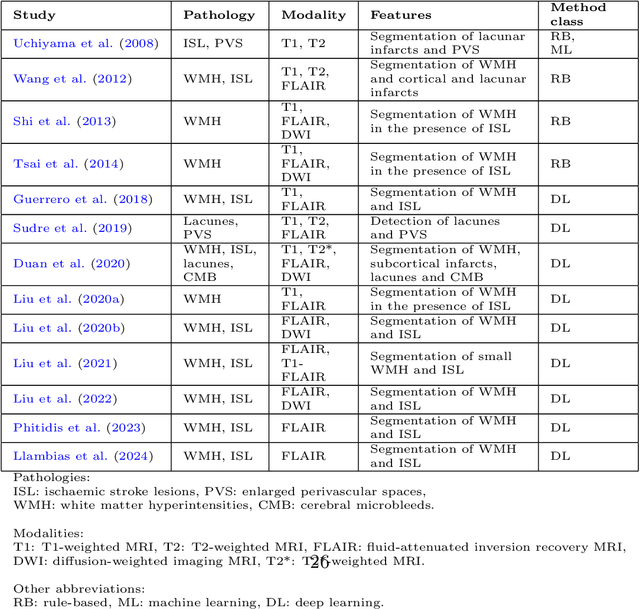
Abstract:Cerebrovascular diseases (CVD) can lead to stroke and dementia. Stroke is the second leading cause of death world wide and dementia incidence is increasing by the year. There are several markers of CVD that are visible on brain imaging, including: white matter hyperintensities (WMH), acute and chronic ischaemic stroke lesions (ISL), lacunes, enlarged perivascular spaces (PVS), acute and chronic haemorrhagic lesions, and cerebral microbleeds (CMB). Brain atrophy also occurs in CVD. These markers are important for patient management and intervention, since they indicate elevated risk of future stroke and dementia. We systematically reviewed automated systems designed to support radiologists reporting on these CVD imaging findings. We considered commercially available software and research publications which identify at least two CVD markers. In total, we included 29 commercial products and 13 research publications. Two distinct types of commercial support system were available: those which identify acute stroke lesions (haemorrhagic and ischaemic) from computed tomography (CT) scans, mainly for the purpose of patient triage; and those which measure WMH and atrophy regionally and longitudinally. In research, WMH and ISL were the markers most frequently analysed together, from magnetic resonance imaging (MRI) scans; lacunes and PVS were each targeted only twice and CMB only once. For stroke, commercially available systems largely support the emergency setting, whilst research systems consider also follow-up and routine scans. The systems to quantify WMH and atrophy are focused on neurodegenerative disease support, where these CVD markers are also of significance. There are currently no openly validated systems, commercially, or in research, performing a comprehensive joint analysis of all CVD markers (WMH, ISL, lacunes, PVS, haemorrhagic lesions, CMB, and atrophy).
Perceptions of Edinburgh: Capturing Neighbourhood Characteristics by Clustering Geoparsed Local News
Sep 17, 2024



Abstract:The communities that we live in affect our health in ways that are complex and hard to define. Moreover, our understanding of the place-based processes affecting health and inequalities is limited. This undermines the development of robust policy interventions to improve local health and well-being. News media provides social and community information that may be useful in health studies. Here we propose a methodology for characterising neighbourhoods by using local news articles. More specifically, we show how we can use Natural Language Processing (NLP) to unlock further information about neighbourhoods by analysing, geoparsing and clustering news articles. Our work is novel because we combine street-level geoparsing tailored to the locality with clustering of full news articles, enabling a more detailed examination of neighbourhood characteristics. We evaluate our outputs and show via a confluence of evidence, both from a qualitative and a quantitative perspective, that the themes we extract from news articles are sensible and reflect many characteristics of the real world. This is significant because it allows us to better understand the effects of neighbourhoods on health. Our findings on neighbourhood characterisation using news data will support a new generation of place-based research which examines a wider set of spatial processes and how they affect health, enabling new epidemiological research.
A Comparative Study on Patient Language across Therapeutic Domains for Effective Patient Voice Classification in Online Health Discussions
Jul 23, 2024Abstract:There exists an invisible barrier between healthcare professionals' perception of a patient's clinical experience and the reality. This barrier may be induced by the environment that hinders patients from sharing their experiences openly with healthcare professionals. As patients are observed to discuss and exchange knowledge more candidly on social media, valuable insights can be leveraged from these platforms. However, the abundance of non-patient posts on social media necessitates filtering out such irrelevant content to distinguish the genuine voices of patients, a task we refer to as patient voice classification. In this study, we analyse the importance of linguistic characteristics in accurately classifying patient voices. Our findings underscore the essential role of linguistic and statistical text similarity analysis in identifying common patterns among patient groups. These results allude to even starker differences in the way patients express themselves at a disease level and across various therapeutic domains. Additionally, we fine-tuned a pre-trained Language Model on the combined datasets with similar linguistic patterns, resulting in a highly accurate automatic patient voice classification. Being the pioneering study on the topic, our focus on extracting authentic patient experiences from social media stands as a crucial step towards advancing healthcare standards and fostering a patient-centric approach.
Infusing clinical knowledge into tokenisers for language models
Jun 20, 2024



Abstract:This study introduces a novel knowledge enhanced tokenisation mechanism, K-Tokeniser, for clinical text processing. Technically, at initialisation stage, K-Tokeniser populates global representations of tokens based on semantic types of domain concepts (such as drugs or diseases) from either a domain ontology like Unified Medical Language System or the training data of the task related corpus. At training or inference stage, sentence level localised context will be utilised for choosing the optimal global token representation to realise the semantic-based tokenisation. To avoid pretraining using the new tokeniser, an embedding initialisation approach is proposed to generate representations for new tokens. Using three transformer-based language models, a comprehensive set of experiments are conducted on four real-world datasets for evaluating K-Tokeniser in a wide range of clinical text analytics tasks including clinical concept and relation extraction, automated clinical coding, clinical phenotype identification, and clinical research article classification. Overall, our models demonstrate consistent improvements over their counterparts in all tasks. In particular, substantial improvements are observed in the automated clinical coding task with 13\% increase on Micro $F_1$ score. Furthermore, K-Tokeniser also shows significant capacities in facilitating quicker converge of language models. Specifically, using K-Tokeniser, the language models would only require 50\% of the training data to achieve the best performance of the baseline tokeniser using all training data in the concept extraction task and less than 20\% of the data for the automated coding task. It is worth mentioning that all these improvements require no pre-training process, making the approach generalisable.
Edinburgh Clinical NLP at MEDIQA-CORR 2024: Guiding Large Language Models with Hints
May 28, 2024Abstract:The MEDIQA-CORR 2024 shared task aims to assess the ability of Large Language Models (LLMs) to identify and correct medical errors in clinical notes. In this study, we evaluate the capability of general LLMs, specifically GPT-3.5 and GPT-4, to identify and correct medical errors with multiple prompting strategies. Recognising the limitation of LLMs in generating accurate corrections only via prompting strategies, we propose incorporating error-span predictions from a smaller, fine-tuned model in two ways: 1) by presenting it as a hint in the prompt and 2) by framing it as multiple-choice questions from which the LLM can choose the best correction. We found that our proposed prompting strategies significantly improve the LLM's ability to generate corrections. Our best-performing solution with 8-shot + CoT + hints ranked sixth in the shared task leaderboard. Additionally, our comprehensive analyses show the impact of the location of the error sentence, the prompted role, and the position of the multiple-choice option on the accuracy of the LLM. This prompts further questions about the readiness of LLM to be implemented in real-world clinical settings.
Edinburgh Clinical NLP at SemEval-2024 Task 2: Fine-tune your model unless you have access to GPT-4
Mar 30, 2024Abstract:The NLI4CT task assesses Natural Language Inference systems in predicting whether hypotheses entail or contradict evidence from Clinical Trial Reports. In this study, we evaluate various Large Language Models (LLMs) with multiple strategies, including Chain-of-Thought, In-Context Learning, and Parameter-Efficient Fine-Tuning (PEFT). We propose a PEFT method to improve the consistency of LLMs by merging adapters that were fine-tuned separately using triplet and language modelling objectives. We found that merging the two PEFT adapters improves the F1 score (+0.0346) and consistency (+0.152) of the LLMs. However, our novel methods did not produce more accurate results than GPT-4 in terms of faithfulness and consistency. Averaging the three metrics, GPT-4 ranks joint-first in the competition with 0.8328. Finally, our contamination analysis with GPT-4 indicates that there was no test data leakage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge