Roma English Owen
A Comparative Study on Patient Language across Therapeutic Domains for Effective Patient Voice Classification in Online Health Discussions
Jul 23, 2024Abstract:There exists an invisible barrier between healthcare professionals' perception of a patient's clinical experience and the reality. This barrier may be induced by the environment that hinders patients from sharing their experiences openly with healthcare professionals. As patients are observed to discuss and exchange knowledge more candidly on social media, valuable insights can be leveraged from these platforms. However, the abundance of non-patient posts on social media necessitates filtering out such irrelevant content to distinguish the genuine voices of patients, a task we refer to as patient voice classification. In this study, we analyse the importance of linguistic characteristics in accurately classifying patient voices. Our findings underscore the essential role of linguistic and statistical text similarity analysis in identifying common patterns among patient groups. These results allude to even starker differences in the way patients express themselves at a disease level and across various therapeutic domains. Additionally, we fine-tuned a pre-trained Language Model on the combined datasets with similar linguistic patterns, resulting in a highly accurate automatic patient voice classification. Being the pioneering study on the topic, our focus on extracting authentic patient experiences from social media stands as a crucial step towards advancing healthcare standards and fostering a patient-centric approach.
Classifying patient voice in social media data using neural networks: A comparison of AI models on different data sources and therapeutic domains
Nov 30, 2023



Abstract:It is essential that healthcare professionals and members of the healthcare community can access and easily understand patient experiences in the real world, so that care standards can be improved and driven towards personalised drug treatment. Social media platforms and message boards are deemed suitable sources of patient experience information, as patients have been observed to discuss and exchange knowledge, look for and provide support online. This paper tests the hypothesis that not all online patient experience information can be treated and collected in the same way, as a result of the inherent differences in the way individuals talk about their journeys, in different therapeutic domains and or data sources. We used linguistic analysis to understand and identify similarities between datasets, across patient language, between data sources (Reddit, SocialGist) and therapeutic domains (cardiovascular, oncology, immunology, neurology). We detected common vocabulary used by patients in the same therapeutic domain across data sources, except for immunology patients, who use unique vocabulary between the two data sources, and compared to all other datasets. We combined linguistically similar datasets to train classifiers (CNN, transformer) to accurately identify patient experience posts from social media, a task we refer to as patient voice classification. The cardiovascular and neurology transformer classifiers perform the best in their respective comparisons for the Reddit data source, achieving F1-scores of 0.865 and 1.0 respectively. The overall best performing classifier is the transformer classifier trained on all data collected for this experiment, achieving F1-scores ranging between 0.863 and 0.995 across all therapeutic domain and data source specific test datasets.
Comparative Analysis of Drug-GPT and ChatGPT LLMs for Healthcare Insights: Evaluating Accuracy and Relevance in Patient and HCP Contexts
Jul 24, 2023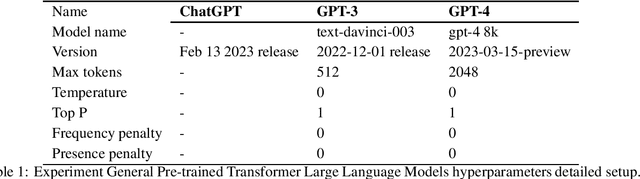
Abstract:This study presents a comparative analysis of three Generative Pre-trained Transformer (GPT) solutions in a question and answer (Q&A) setting: Drug-GPT 3, Drug-GPT 4, and ChatGPT, in the context of healthcare applications. The objective is to determine which model delivers the most accurate and relevant information in response to prompts related to patient experiences with atopic dermatitis (AD) and healthcare professional (HCP) discussions about diabetes. The results demonstrate that while all three models are capable of generating relevant and accurate responses, Drug-GPT 3 and Drug-GPT 4, which are supported by curated datasets of patient and HCP social media and message board posts, provide more targeted and in-depth insights. ChatGPT, a more general-purpose model, generates broader and more general responses, which may be valuable for readers seeking a high-level understanding of the topics but may lack the depth and personal insights found in the answers generated by the specialized Drug-GPT models. This comparative analysis highlights the importance of considering the language model's perspective, depth of knowledge, and currency when evaluating the usefulness of generated information in healthcare applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge