Huayu Zhang
Everything-Grasping (EG) Gripper: A Universal Gripper with Synergistic Suction-Grasping Capabilities for Cross-Scale and Cross-State Manipulation
Oct 06, 2025Abstract:Grasping objects across vastly different sizes and physical states-including both solids and liquids-with a single robotic gripper remains a fundamental challenge in soft robotics. We present the Everything-Grasping (EG) Gripper, a soft end-effector that synergistically integrates distributed surface suction with internal granular jamming, enabling cross-scale and cross-state manipulation without requiring airtight sealing at the contact interface with target objects. The EG Gripper can handle objects with surface areas ranging from sub-millimeter scale 0.2 mm2 (glass bead) to over 62,000 mm2 (A4 sized paper and woven bag), enabling manipulation of objects nearly 3,500X smaller and 88X larger than its own contact area (approximated at 707 mm2 for a 30 mm-diameter base). We further introduce a tactile sensing framework that combines liquid detection and pressure-based suction feedback, enabling real-time differentiation between solid and liquid targets. Guided by the actile-Inferred Grasping Mode Selection (TIGMS) algorithm, the gripper autonomously selects grasping modes based on distributed pressure and voltage signals. Experiments across diverse tasks-including underwater grasping, fragile object handling, and liquid capture-demonstrate robust and repeatable performance. To our knowledge, this is the first soft gripper to reliably grasp both solid and liquid objects across scales using a unified compliant architecture.
FairHuman: Boosting Hand and Face Quality in Human Image Generation with Minimum Potential Delay Fairness in Diffusion Models
Jul 03, 2025Abstract:Image generation has achieved remarkable progress with the development of large-scale text-to-image models, especially diffusion-based models. However, generating human images with plausible details, such as faces or hands, remains challenging due to insufficient supervision of local regions during training. To address this issue, we propose FairHuman, a multi-objective fine-tuning approach designed to enhance both global and local generation quality fairly. Specifically, we first construct three learning objectives: a global objective derived from the default diffusion objective function and two local objectives for hands and faces based on pre-annotated positional priors. Subsequently, we derive the optimal parameter updating strategy under the guidance of the Minimum Potential Delay (MPD) criterion, thereby attaining fairness-ware optimization for this multi-objective problem. Based on this, our proposed method can achieve significant improvements in generating challenging local details while maintaining overall quality. Extensive experiments showcase the effectiveness of our method in improving the performance of human image generation under different scenarios.
Adverse Event Extraction from Discharge Summaries: A New Dataset, Annotation Scheme, and Initial Findings
Jun 17, 2025Abstract:In this work, we present a manually annotated corpus for Adverse Event (AE) extraction from discharge summaries of elderly patients, a population often underrepresented in clinical NLP resources. The dataset includes 14 clinically significant AEs-such as falls, delirium, and intracranial haemorrhage, along with contextual attributes like negation, diagnosis type, and in-hospital occurrence. Uniquely, the annotation schema supports both discontinuous and overlapping entities, addressing challenges rarely tackled in prior work. We evaluate multiple models using FlairNLP across three annotation granularities: fine-grained, coarse-grained, and coarse-grained with negation. While transformer-based models (e.g., BERT-cased) achieve strong performance on document-level coarse-grained extraction (F1 = 0.943), performance drops notably for fine-grained entity-level tasks (e.g., F1 = 0.675), particularly for rare events and complex attributes. These results demonstrate that despite high-level scores, significant challenges remain in detecting underrepresented AEs and capturing nuanced clinical language. Developed within a Trusted Research Environment (TRE), the dataset is available upon request via DataLoch and serves as a robust benchmark for evaluating AE extraction methods and supporting future cross-dataset generalisation.
MP-Mat: A 3D-and-Instance-Aware Human Matting and Editing Framework with Multiplane Representation
Apr 20, 2025

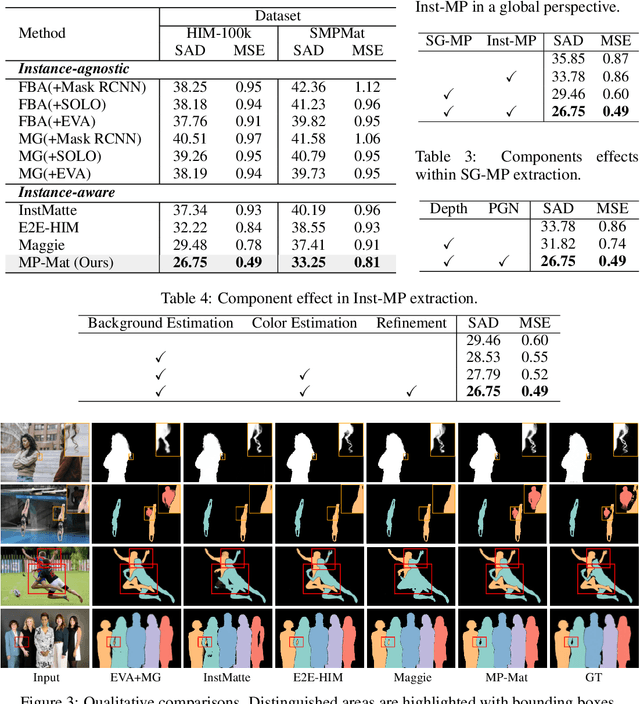

Abstract:Human instance matting aims to estimate an alpha matte for each human instance in an image, which is challenging as it easily fails in complex cases requiring disentangling mingled pixels belonging to multiple instances along hairy and thin boundary structures. In this work, we address this by introducing MP-Mat, a novel 3D-and-instance-aware matting framework with multiplane representation, where the multiplane concept is designed from two different perspectives: scene geometry level and instance level. Specifically, we first build feature-level multiplane representations to split the scene into multiple planes based on depth differences. This approach makes the scene representation 3D-aware, and can serve as an effective clue for splitting instances in different 3D positions, thereby improving interpretability and boundary handling ability especially in occlusion areas. Then, we introduce another multiplane representation that splits the scene in an instance-level perspective, and represents each instance with both matte and color. We also treat background as a special instance, which is often overlooked by existing methods. Such an instance-level representation facilitates both foreground and background content awareness, and is useful for other down-stream tasks like image editing. Once built, the representation can be reused to realize controllable instance-level image editing with high efficiency. Extensive experiments validate the clear advantage of MP-Mat in matting task. We also demonstrate its superiority in image editing tasks, an area under-explored by existing matting-focused methods, where our approach under zero-shot inference even outperforms trained specialized image editing techniques by large margins. Code is open-sourced at https://github.com/JiaoSiyi/MPMat.git}.
Object-Aware Video Matting with Cross-Frame Guidance
Mar 03, 2025



Abstract:Recently, trimap-free methods have drawn increasing attention in human video matting due to their promising performance. Nevertheless, these methods still suffer from the lack of deterministic foreground-background cues, which impairs their ability to consistently identify and locate foreground targets over time and mine fine-grained details. In this paper, we present a trimap-free Object-Aware Video Matting (OAVM) framework, which can perceive different objects, enabling joint recognition of foreground objects and refinement of edge details. Specifically, we propose an Object-Guided Correction and Refinement (OGCR) module, which employs cross-frame guidance to aggregate object-level instance information into pixel-level detail features, thereby promoting their synergy. Furthermore, we design a Sequential Foreground Merging augmentation strategy to diversify sequential scenarios and enhance capacity of the network for object discrimination. Extensive experiments on recent widely used synthetic and real-world benchmarks demonstrate the state-of-the-art performance of our OAVM with only an initial coarse mask. The code and model will be available.
A CT-guided Control Framework of a Robotic Flexible Endoscope for the Diagnosis of the Maxillary Sinusitis
Oct 27, 2024



Abstract:Flexible endoscopes are commonly adopted in narrow and confined anatomical cavities due to their higher reachability and dexterity. However, prolonged and unintuitive manipulation of these endoscopes leads to an increased workload on surgeons and risks of collision. To address these challenges, this paper proposes a CT-guided control framework for the diagnosis of maxillary sinusitis by using a robotic flexible endoscope. In the CT-guided control framework, a feasible path to the target position in the maxillary sinus cavity for the robotic flexible endoscope is designed. Besides, an optimal control scheme is proposed to autonomously control the robotic flexible endoscope to follow the feasible path. This greatly improves the efficiency and reduces the workload for surgeons. Several experiments were conducted based on a widely utilized sinus phantom, and the results showed that the robotic flexible endoscope can accurately and autonomously follow the feasible path and reach the target position in the maxillary sinus cavity. The results also verified the feasibility of the CT-guided control framework, which contributes an effective approach to early diagnosis of sinusitis in the future.
Infusing clinical knowledge into tokenisers for language models
Jun 20, 2024



Abstract:This study introduces a novel knowledge enhanced tokenisation mechanism, K-Tokeniser, for clinical text processing. Technically, at initialisation stage, K-Tokeniser populates global representations of tokens based on semantic types of domain concepts (such as drugs or diseases) from either a domain ontology like Unified Medical Language System or the training data of the task related corpus. At training or inference stage, sentence level localised context will be utilised for choosing the optimal global token representation to realise the semantic-based tokenisation. To avoid pretraining using the new tokeniser, an embedding initialisation approach is proposed to generate representations for new tokens. Using three transformer-based language models, a comprehensive set of experiments are conducted on four real-world datasets for evaluating K-Tokeniser in a wide range of clinical text analytics tasks including clinical concept and relation extraction, automated clinical coding, clinical phenotype identification, and clinical research article classification. Overall, our models demonstrate consistent improvements over their counterparts in all tasks. In particular, substantial improvements are observed in the automated clinical coding task with 13\% increase on Micro $F_1$ score. Furthermore, K-Tokeniser also shows significant capacities in facilitating quicker converge of language models. Specifically, using K-Tokeniser, the language models would only require 50\% of the training data to achieve the best performance of the baseline tokeniser using all training data in the concept extraction task and less than 20\% of the data for the automated coding task. It is worth mentioning that all these improvements require no pre-training process, making the approach generalisable.
Explainable Bayesian Recurrent Neural Smoother to Capture Global State Evolutionary Correlations
Jun 17, 2024Abstract:Through integrating the evolutionary correlations across global states in the bidirectional recursion, an explainable Bayesian recurrent neural smoother (EBRNS) is proposed for offline data-assisted fixed-interval state smoothing. At first, the proposed model, containing global states in the evolutionary interval, is transformed into an equivalent model with bidirectional memory. This transformation incorporates crucial global state information with support for bi-directional recursive computation. For the transformed model, the joint state-memory-trend Bayesian filtering and smoothing frameworks are derived by introducing the bidirectional memory iteration mechanism and offline data into Bayesian estimation theory. The derived frameworks are implemented using the Gaussian approximation to ensure analytical properties and computational efficiency. Finally, the neural network modules within EBRNS and its two-stage training scheme are designed. Unlike most existing approaches that artificially combine deep learning and model-based estimation, the bidirectional recursion and internal gated structures of EBRNS are naturally derived from Bayesian estimation theory, explainably integrating prior model knowledge, online measurement, and offline data. Experiments on representative real-world datasets demonstrate that the high smoothing accuracy of EBRNS is accompanied by data efficiency and a lightweight parameter scale.
Deterministic Computing Power Networking: Architecture, Technologies and Prospects
Jan 31, 2024Abstract:With the development of new Internet services such as computation-intensive and delay-sensitive tasks, the traditional "Best Effort" network transmission mode has been greatly challenged. The network system is urgently required to provide end-to-end transmission determinacy and computing determinacy for new applications to ensure the safe and efficient operation of services. Based on the research of the convergence of computing and networking, a new network paradigm named deterministic computing power networking (Det-CPN) is proposed. In this article, we firstly introduce the research advance of computing power networking. And then the motivations and scenarios of Det-CPN are analyzed. Following that, we present the system architecture, technological capabilities, workflow as well as key technologies for Det-CPN. Finally, the challenges and future trends of Det-CPN are analyzed and discussed.
TIA: A Teaching Intonation Assessment Dataset in Real Teaching Situations
Dec 14, 2023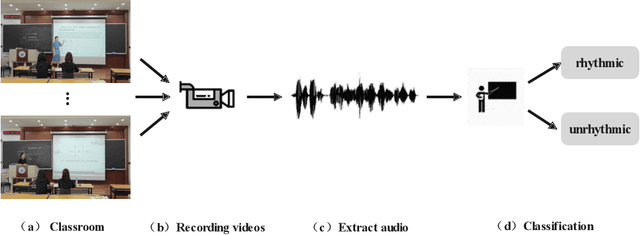
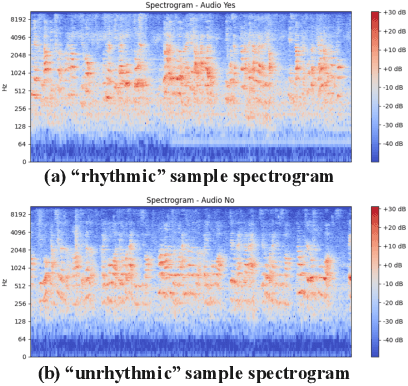
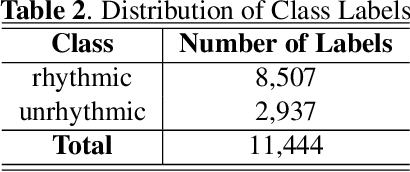
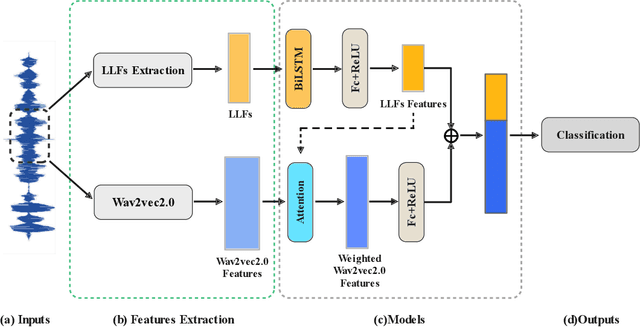
Abstract:Intonation is one of the important factors affecting the teaching language arts, so it is an urgent problem to be addressed by evaluating the teachers' intonation through artificial intelligence technology. However, the lack of an intonation assessment dataset has hindered the development of the field. To this end, this paper constructs a Teaching Intonation Assessment (TIA) dataset for the first time in real teaching situations. This dataset covers 9 disciplines, 396 teachers, total of 11,444 utterance samples with a length of 15 seconds. In order to test the validity of the dataset, this paper proposes a teaching intonation assessment model (TIAM) based on low-level and deep-level features of speech. The experimental results show that TIAM based on the dataset constructed in this paper is basically consistent with the results of manual evaluation, and the results are better than the baseline models, which proves the effectiveness of the evaluation model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge