Víctor Suárez-Paniagua
Combining Denoising Autoencoders with Contrastive Learning to fine-tune Transformer Models
May 23, 2024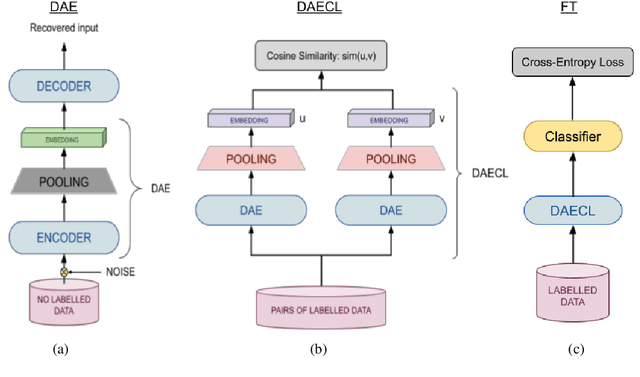
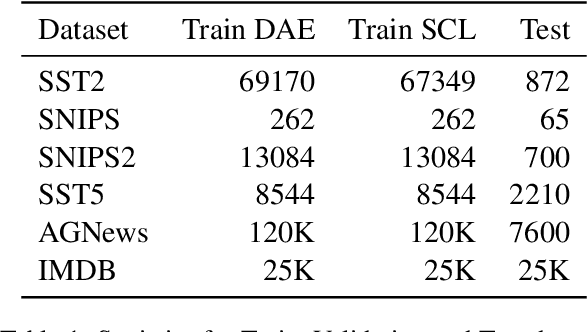
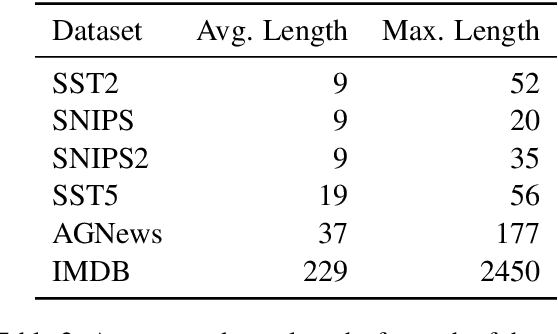
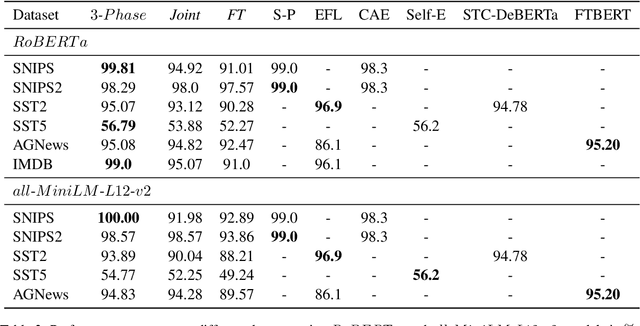
Abstract:Recently, using large pretrained Transformer models for transfer learning tasks has evolved to the point where they have become one of the flagship trends in the Natural Language Processing (NLP) community, giving rise to various outlooks such as prompt-based, adapters or combinations with unsupervised approaches, among many others. This work proposes a 3 Phase technique to adjust a base model for a classification task. First, we adapt the model's signal to the data distribution by performing further training with a Denoising Autoencoder (DAE). Second, we adjust the representation space of the output to the corresponding classes by clustering through a Contrastive Learning (CL) method. In addition, we introduce a new data augmentation approach for Supervised Contrastive Learning to correct the unbalanced datasets. Third, we apply fine-tuning to delimit the predefined categories. These different phases provide relevant and complementary knowledge to the model to learn the final task. We supply extensive experimental results on several datasets to demonstrate these claims. Moreover, we include an ablation study and compare the proposed method against other ways of combining these techniques.
* 1 figure, 7 tables, 12 pages
Multilingual News Location Detection using an Entity-Based Siamese Network with Semi-Supervised Contrastive Learning and Knowledge Base
Dec 22, 2022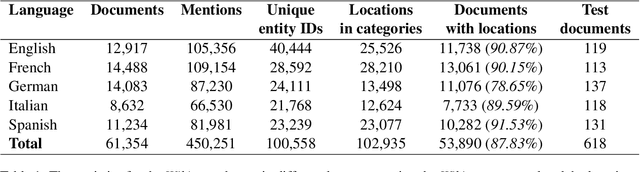
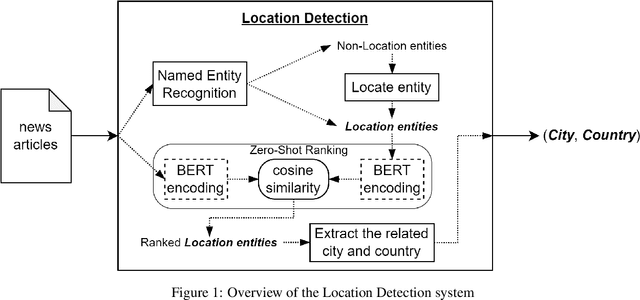
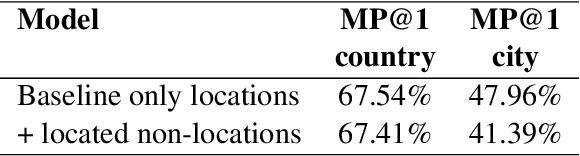
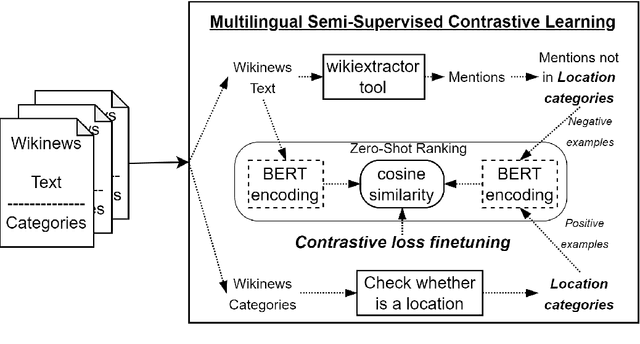
Abstract:Early detection of relevant locations in a piece of news is especially important in extreme events such as environmental disasters, war conflicts, disease outbreaks, or political turmoils. Additionally, this detection also helps recommender systems to promote relevant news based on user locations. Note that, when the relevant locations are not mentioned explicitly in the text, state-of-the-art methods typically fail to recognize them because these methods rely on syntactic recognition. In contrast, by incorporating a knowledge base and connecting entities with their locations, our system successfully infers the relevant locations even when they are not mentioned explicitly in the text. To evaluate the effectiveness of our approach, and due to the lack of datasets in this area, we also contribute to the research community with a gold-standard multilingual news-location dataset, NewsLOC. It contains the annotation of the relevant locations (and their WikiData IDs) of 600+ Wikinews articles in five different languages: English, French, German, Italian, and Spanish. Through experimental evaluations, we show that our proposed system outperforms the baselines and the fine-tuned version of the model using semi-supervised data that increases the classification rate. The source code and the NewsLOC dataset are publicly available for being used by the research community at https://github.com/vsuarezpaniagua/NewsLocation.
Ontology-Based and Weakly Supervised Rare Disease Phenotyping from Clinical Notes
May 11, 2022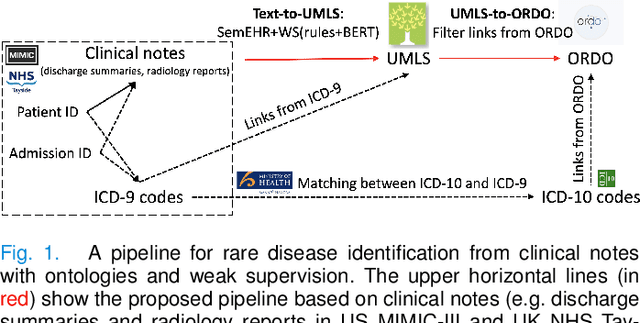
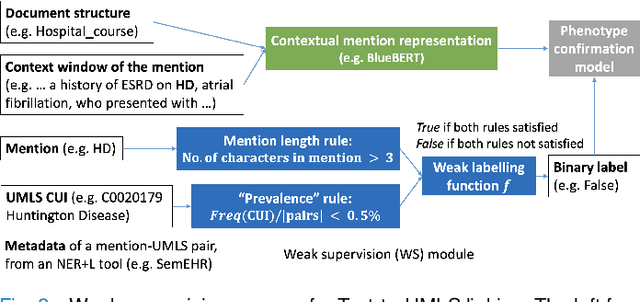
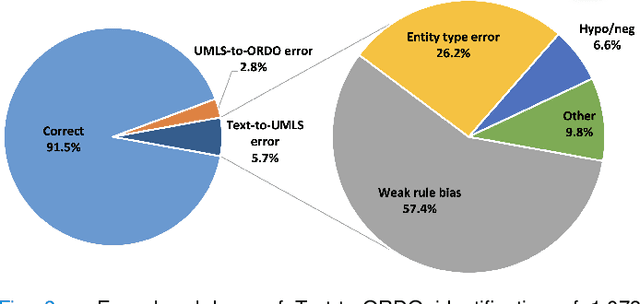
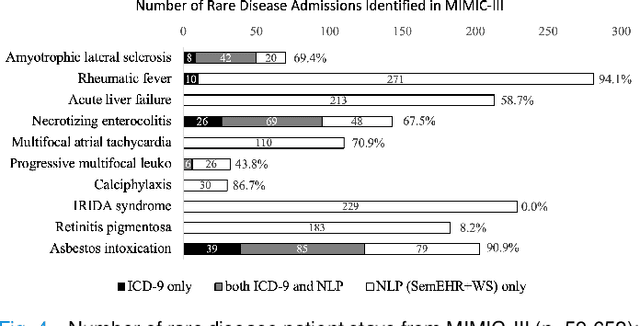
Abstract:Computational text phenotyping is the practice of identifying patients with certain disorders and traits from clinical notes. Rare diseases are challenging to be identified due to few cases available for machine learning and the need for data annotation from domain experts. We propose a method using ontologies and weak supervision, with recent pre-trained contextual representations from Bi-directional Transformers (e.g. BERT). The ontology-based framework includes two steps: (i) Text-to-UMLS, extracting phenotypes by contextually linking mentions to concepts in Unified Medical Language System (UMLS), with a Named Entity Recognition and Linking (NER+L) tool, SemEHR, and weak supervision with customised rules and contextual mention representation; (ii) UMLS-to-ORDO, matching UMLS concepts to rare diseases in Orphanet Rare Disease Ontology (ORDO). The weakly supervised approach is proposed to learn a phenotype confirmation model to improve Text-to-UMLS linking, without annotated data from domain experts. We evaluated the approach on three clinical datasets of discharge summaries and radiology reports from two institutions in the US and the UK. Our best weakly supervised method achieved 81.4% precision and 91.4% recall on extracting rare disease UMLS phenotypes from MIMIC-III discharge summaries. The overall pipeline processing clinical notes can surface rare disease cases, mostly uncaptured in structured data (manually assigned ICD codes). Results on radiology reports from MIMIC-III and NHS Tayside were consistent with the discharge summaries. We discuss the usefulness of the weak supervision approach and propose directions for future studies.
Rare Disease Identification from Clinical Notes with Ontologies and Weak Supervision
May 08, 2021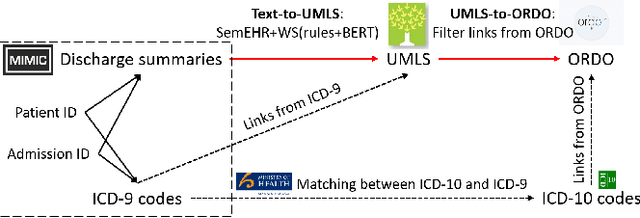
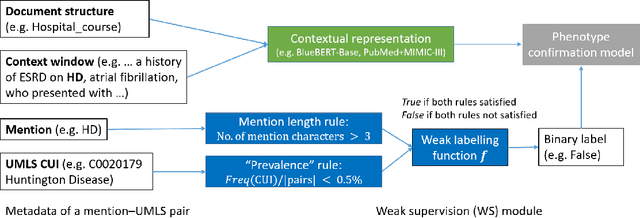


Abstract:The identification of rare diseases from clinical notes with Natural Language Processing (NLP) is challenging due to the few cases available for machine learning and the need of data annotation from clinical experts. We propose a method using ontologies and weak supervision. The approach includes two steps: (i) Text-to-UMLS, linking text mentions to concepts in Unified Medical Language System (UMLS), with a named entity linking tool (e.g. SemEHR) and weak supervision based on customised rules and Bidirectional Encoder Representations from Transformers (BERT) based contextual representations, and (ii) UMLS-to-ORDO, matching UMLS concepts to rare diseases in Orphanet Rare Disease Ontology (ORDO). Using MIMIC-III discharge summaries as a case study, we show that the Text-to-UMLS process can be greatly improved with weak supervision, without any annotated data from domain experts. Our analysis shows that the overall pipeline processing discharge summaries can surface rare disease cases, which are mostly uncaptured in manual ICD codes of the hospital admissions.
A Systematic Review of Natural Language Processing Applied to Radiology Reports
Feb 18, 2021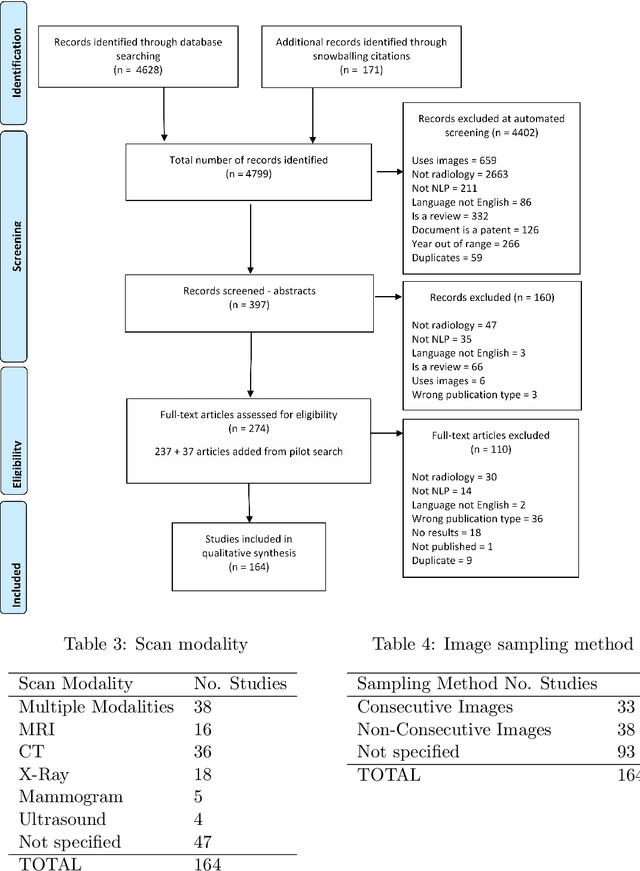
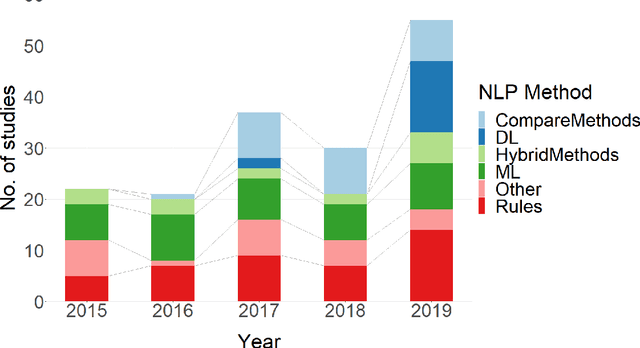
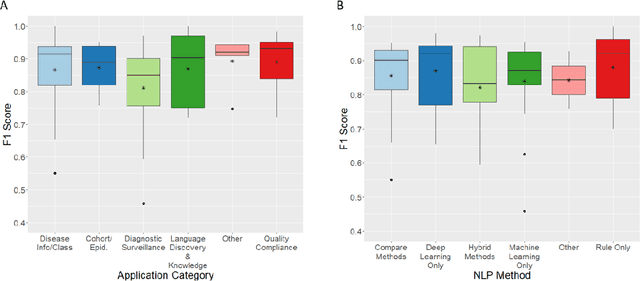
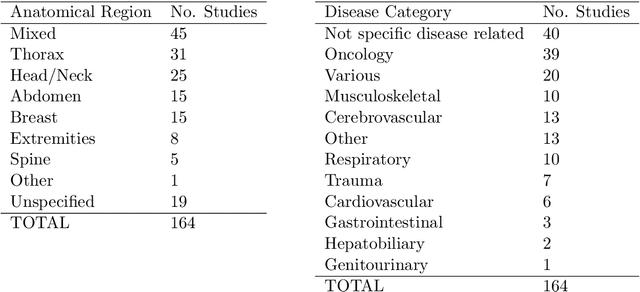
Abstract:NLP has a significant role in advancing healthcare and has been found to be key in extracting structured information from radiology reports. Understanding recent developments in NLP application to radiology is of significance but recent reviews on this are limited. This study systematically assesses recent literature in NLP applied to radiology reports. Our automated literature search yields 4,799 results using automated filtering, metadata enriching steps and citation search combined with manual review. Our analysis is based on 21 variables including radiology characteristics, NLP methodology, performance, study, and clinical application characteristics. We present a comprehensive analysis of the 164 publications retrieved with each categorised into one of 6 clinical application categories. Deep learning use increases but conventional machine learning approaches are still prevalent. Deep learning remains challenged when data is scarce and there is little evidence of adoption into clinical practice. Despite 17% of studies reporting greater than 0.85 F1 scores, it is hard to comparatively evaluate these approaches given that most of them use different datasets. Only 14 studies made their data and 15 their code available with 10 externally validating results. Automated understanding of clinical narratives of the radiology reports has the potential to enhance the healthcare process but reproducibility and explainability of models are important if the domain is to move applications into clinical use. More could be done to share code enabling validation of methods on different institutional data and to reduce heterogeneity in reporting of study properties allowing inter-study comparisons. Our results have significance for researchers providing a systematic synthesis of existing work to build on, identify gaps, opportunities for collaboration and avoid duplication.
Explainable Automated Coding of Clinical Notes using Hierarchical Label-wise Attention Networks and Label Embedding Initialisation
Oct 30, 2020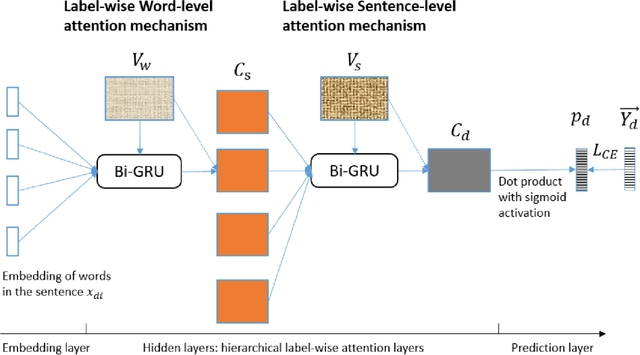
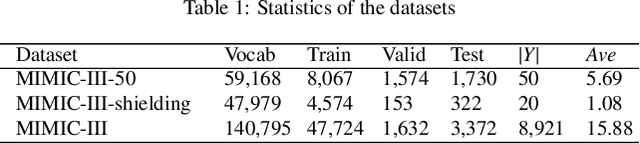
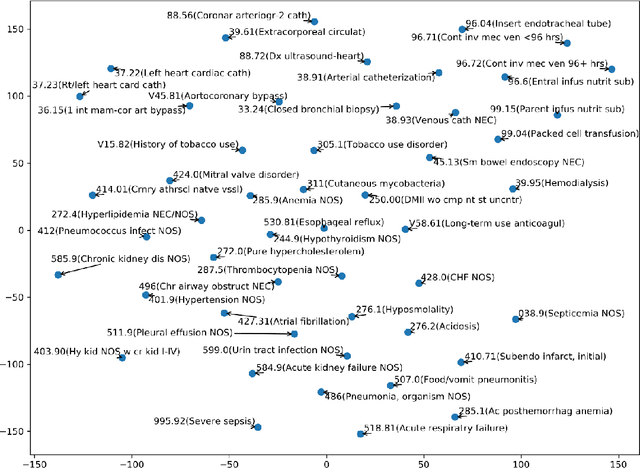
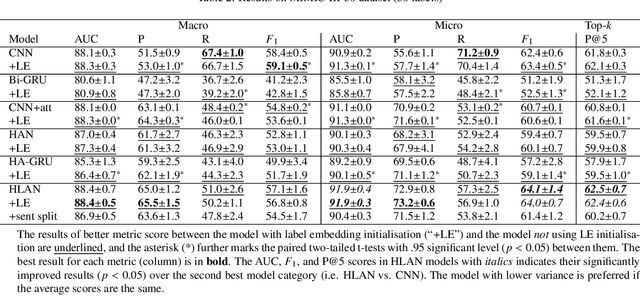
Abstract:Diagnostic or procedural coding of clinical notes aims to derive a coded summary of disease-related information about patients. Such coding is usually done manually in hospitals but could potentially be automated to improve the efficiency and accuracy of medical coding. Recent studies on deep learning for automated medical coding achieved promising performances. However, the explainability of these models is usually poor, preventing them to be used confidently in supporting clinical practice. Another limitation is that these models mostly assume independence among labels, ignoring the complex correlation among medical codes which can potentially be exploited to improve the performance. We propose a Hierarchical Label-wise Attention Network (HLAN), which aimed to interpret the model by quantifying importance (as attention weights) of words and sentences related to each of the labels. Secondly, we propose to enhance the major deep learning models with a label embedding (LE) initialisation approach, which learns a dense, continuous vector representation and then injects the representation into the final layers and the label-wise attention layers in the models. We evaluated the methods using three settings on the MIMIC-III discharge summaries: full codes, top-50 codes, and the UK NHS COVID-19 shielding codes. Experiments were conducted to compare HLAN and LE initialisation to the state-of-the-art neural network based methods. HLAN achieved the best Micro-level AUC and $F_1$ on the top-50 code prediction and comparable results on the NHS COVID-19 shielding code prediction to other models. By highlighting the most salient words and sentences for each label, HLAN showed more meaningful and comprehensive model interpretation compared to its downgraded baselines and the CNN-based models. LE initialisation consistently boosted most deep learning models for automated medical coding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge