Emma Whitfield
Rare Disease Identification from Clinical Notes with Ontologies and Weak Supervision
May 08, 2021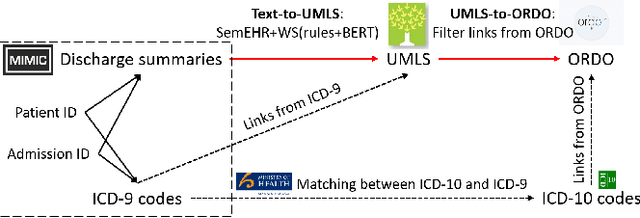
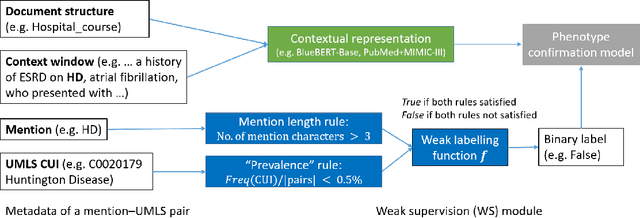


Abstract:The identification of rare diseases from clinical notes with Natural Language Processing (NLP) is challenging due to the few cases available for machine learning and the need of data annotation from clinical experts. We propose a method using ontologies and weak supervision. The approach includes two steps: (i) Text-to-UMLS, linking text mentions to concepts in Unified Medical Language System (UMLS), with a named entity linking tool (e.g. SemEHR) and weak supervision based on customised rules and Bidirectional Encoder Representations from Transformers (BERT) based contextual representations, and (ii) UMLS-to-ORDO, matching UMLS concepts to rare diseases in Orphanet Rare Disease Ontology (ORDO). Using MIMIC-III discharge summaries as a case study, we show that the Text-to-UMLS process can be greatly improved with weak supervision, without any annotated data from domain experts. Our analysis shows that the overall pipeline processing discharge summaries can surface rare disease cases, which are mostly uncaptured in manual ICD codes of the hospital admissions.
Bayesian Screening: Multi-test Bayesian Optimization Applied to in silico Material Screening
Sep 11, 2020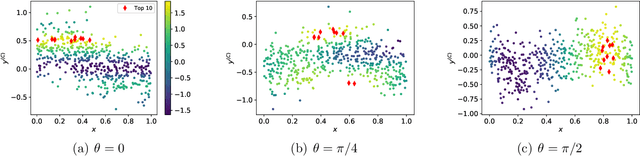
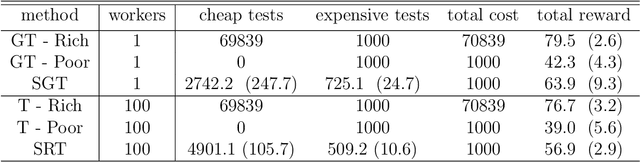
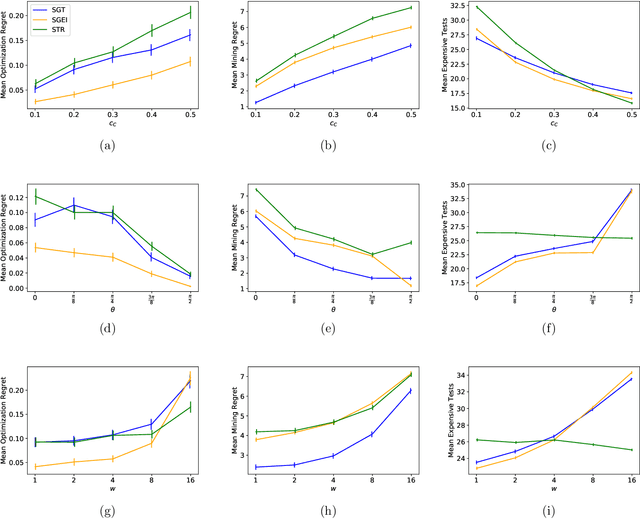
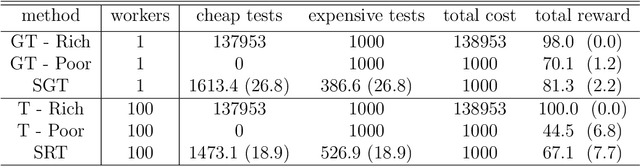
Abstract:We present new multi-test Bayesian optimization models and algorithms for use in large scale material screening applications. Our screening problems are designed around two tests, one expensive and one cheap. This paper differs from other recent work on multi-test Bayesian optimization through use of a flexible model that allows for complex, non-linear relationships between the cheap and expensive test scores. This additional modeling flexibility is essential in the material screening applications which we describe. We demonstrate the power of our new algorithms on a family of synthetic toy problems as well as on real data from two large scale screening studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge