Peiyu Duan
Causal Modeling of fMRI Time-series for Interpretable Autism Spectrum Disorder Classification
Feb 21, 2025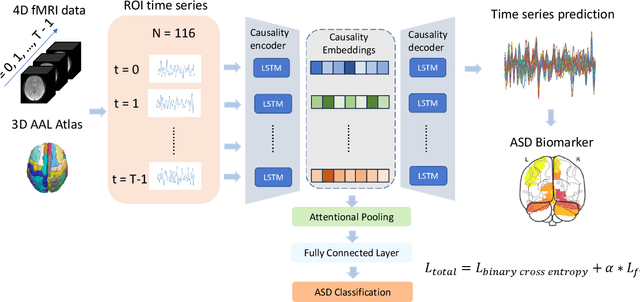
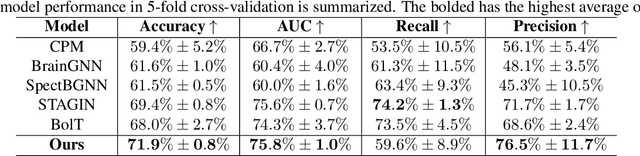
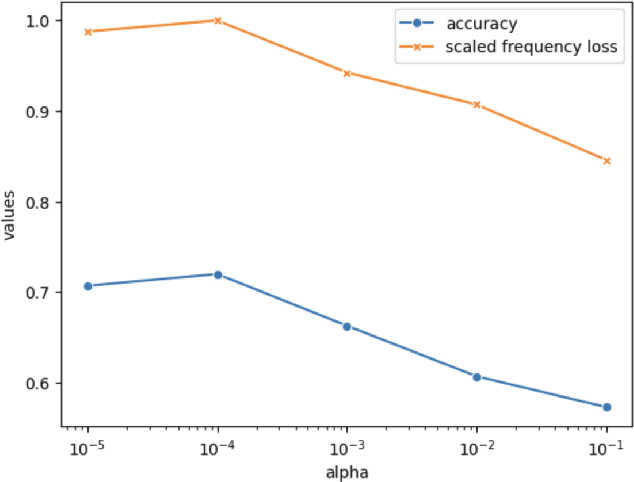
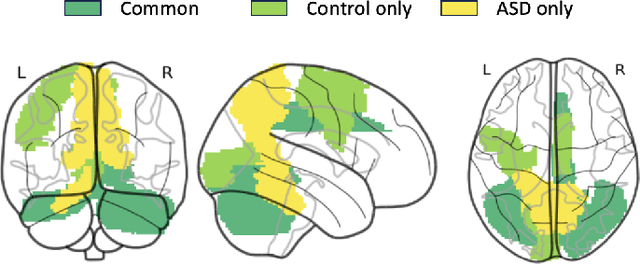
Abstract:Autism spectrum disorder (ASD) is a neurological and developmental disorder that affects social and communicative behaviors. It emerges in early life and is generally associated with lifelong disabilities. Thus, accurate and early diagnosis could facilitate treatment outcomes for those with ASD. Functional magnetic resonance imaging (fMRI) is a useful tool that measures changes in brain signaling to facilitate our understanding of ASD. Much effort is being made to identify ASD biomarkers using various connectome-based machine learning and deep learning classifiers. However, correlation-based models cannot capture the non-linear interactions between brain regions. To solve this problem, we introduce a causality-inspired deep learning model that uses time-series information from fMRI and captures causality among ROIs useful for ASD classification. The model is compared with other baseline and state-of-the-art models with 5-fold cross-validation on the ABIDE dataset. We filtered the dataset by choosing all the images with mean FD less than 15mm to ensure data quality. Our proposed model achieved the highest average classification accuracy of 71.9% and an average AUC of 75.8%. Moreover, the inter-ROI causality interpretation of the model suggests that the left precuneus, right precuneus, and cerebellum are placed in the top 10 ROIs in inter-ROI causality among the ASD population. In contrast, these ROIs are not ranked in the top 10 in the control population. We have validated our findings with the literature and found that abnormalities in these ROIs are often associated with ASD.
STNAGNN: Spatiotemporal Node Attention Graph Neural Network for Task-based fMRI Analysis
Jun 17, 2024Abstract:Task-based fMRI uses actions or stimuli to trigger task-specific brain responses and measures them using BOLD contrast. Despite the significant task-induced spatiotemporal brain activation fluctuations, most studies on task-based fMRI ignore the task context information aligned with fMRI and consider task-based fMRI a coherent sequence. In this paper, we show that using the task structures as data-driven guidance is effective for spatiotemporal analysis. We propose STNAGNN, a GNN-based spatiotemporal architecture, and validate its performance in an autism classification task. The trained model is also interpreted for identifying autism-related spatiotemporal brain biomarkers.
Efficient Annotation for Medical Image Analysis: A One-Pass Selective Annotation Approach
Sep 15, 2023



Abstract:Annotating biomedical images for supervised learning is a complex and labor-intensive task due to data diversity and its intricate nature. In this paper, we propose an innovative method, the efficient one-pass selective annotation (EPOSA), that significantly reduces the annotation burden while maintaining robust model performance. Our approach employs a variational autoencoder (VAE) to extract salient features from unannotated images, which are subsequently clustered using the DBSCAN algorithm. This process groups similar images together, forming distinct clusters. We then use a two-stage sample selection algorithm, called representative selection (RepSel), to form a selected dataset. The first stage is a Markov Chain Monte Carlo (MCMC) sampling technique to select representative samples from each cluster for annotations. This selection process is the second stage, which is guided by the principle of maximizing intra-cluster mutual information and minimizing inter-cluster mutual information. This ensures a diverse set of features for model training and minimizes outlier inclusion. The selected samples are used to train a VGG-16 network for image classification. Experimental results on the Med-MNIST dataset demonstrate that our proposed EPOSA outperforms random selection and other state-of-the-art methods under the same annotation budget, presenting a promising direction for efficient and effective annotation in medical image analysis.
Rapid Brain Meninges Surface Reconstruction with Layer Topology Guarantee
Apr 13, 2023



Abstract:The meninges, located between the skull and brain, are composed of three membrane layers: the pia, the arachnoid, and the dura. Reconstruction of these layers can aid in studying volume differences between patients with neurodegenerative diseases and normal aging subjects. In this work, we use convolutional neural networks (CNNs) to reconstruct surfaces representing meningeal layer boundaries from magnetic resonance (MR) images. We first use the CNNs to predict the signed distance functions (SDFs) representing these surfaces while preserving their anatomical ordering. The marching cubes algorithm is then used to generate continuous surface representations; both the subarachnoid space (SAS) and the intracranial volume (ICV) are computed from these surfaces. The proposed method is compared to a state-of-the-art deformable model-based reconstruction method, and we show that our method can reconstruct smoother and more accurate surfaces using less computation time. Finally, we conduct experiments with volumetric analysis on both subjects with multiple sclerosis and healthy controls. For healthy and MS subjects, ICVs and SAS volumes are found to be significantly correlated to sex (p<0.01) and age (p<0.03) changes, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge