Ahmed Alshareef
Brightness-Invariant Tracking Estimation in Tagged MRI
May 23, 2025Abstract:Magnetic resonance (MR) tagging is an imaging technique for noninvasively tracking tissue motion in vivo by creating a visible pattern of magnetization saturation (tags) that deforms with the tissue. Due to longitudinal relaxation and progression to steady-state, the tags and tissue brightnesses change over time, which makes tracking with optical flow methods error-prone. Although Fourier methods can alleviate these problems, they are also sensitive to brightness changes as well as spectral spreading due to motion. To address these problems, we introduce the brightness-invariant tracking estimation (BRITE) technique for tagged MRI. BRITE disentangles the anatomy from the tag pattern in the observed tagged image sequence and simultaneously estimates the Lagrangian motion. The inherent ill-posedness of this problem is addressed by leveraging the expressive power of denoising diffusion probabilistic models to represent the probabilistic distribution of the underlying anatomy and the flexibility of physics-informed neural networks to estimate biologically-plausible motion. A set of tagged MR images of a gel phantom was acquired with various tag periods and imaging flip angles to demonstrate the impact of brightness variations and to validate our method. The results show that BRITE achieves more accurate motion and strain estimates as compared to other state of the art methods, while also being resistant to tag fading.
Is Registering Raw Tagged-MR Enough for Strain Estimation in the Era of Deep Learning?
Jan 31, 2024Abstract:Magnetic Resonance Imaging with tagging (tMRI) has long been utilized for quantifying tissue motion and strain during deformation. However, a phenomenon known as tag fading, a gradual decrease in tag visibility over time, often complicates post-processing. The first contribution of this study is to model tag fading by considering the interplay between $T_1$ relaxation and the repeated application of radio frequency (RF) pulses during serial imaging sequences. This is a factor that has been overlooked in prior research on tMRI post-processing. Further, we have observed an emerging trend of utilizing raw tagged MRI within a deep learning-based (DL) registration framework for motion estimation. In this work, we evaluate and analyze the impact of commonly used image similarity objectives in training DL registrations on raw tMRI. This is then compared with the Harmonic Phase-based approach, a traditional approach which is claimed to be robust to tag fading. Our findings, derived from both simulated images and an actual phantom scan, reveal the limitations of various similarity losses in raw tMRI and emphasize caution in registration tasks where image intensity changes over time.
Data-driven Uncertainty Quantification in Computational Human Head Models
Oct 29, 2021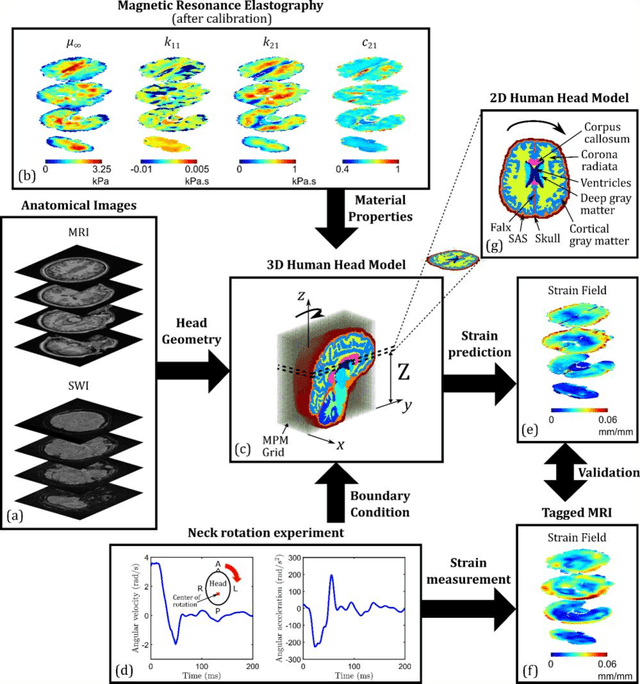
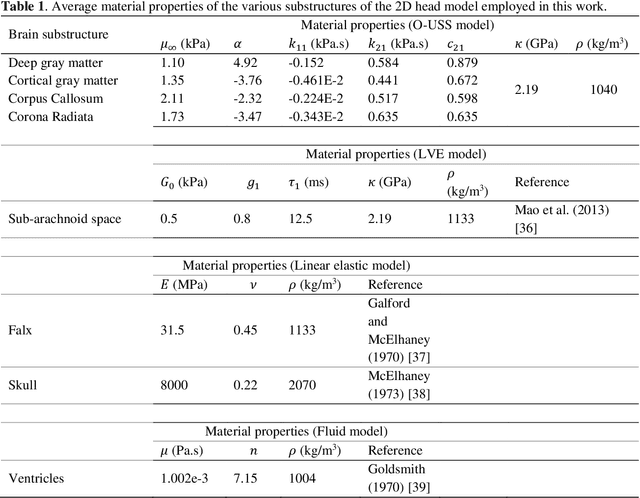
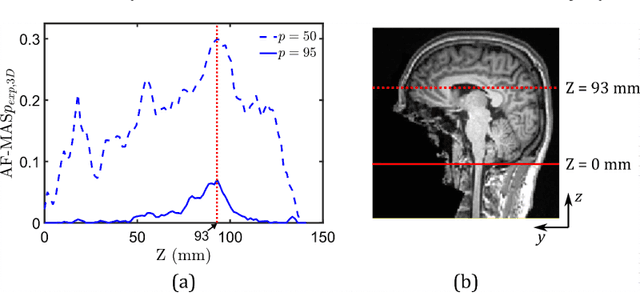
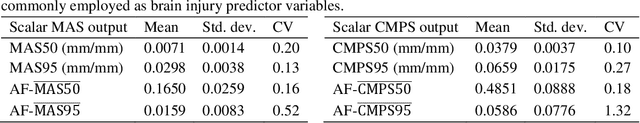
Abstract:Computational models of the human head are promising tools for estimating the impact-induced response of brain, and thus play an important role in the prediction of traumatic brain injury. Modern biofidelic head model simulations are associated with very high computational cost, and high-dimensional inputs and outputs, which limits the applicability of traditional uncertainty quantification (UQ) methods on these systems. In this study, a two-stage, data-driven manifold learning-based framework is proposed for UQ of computational head models. This framework is demonstrated on a 2D subject-specific head model, where the goal is to quantify uncertainty in the simulated strain fields (i.e., output), given variability in the material properties of different brain substructures (i.e., input). In the first stage, a data-driven method based on multi-dimensional Gaussian kernel-density estimation and diffusion maps is used to generate realizations of the input random vector directly from the available data. Computational simulations of a small number of realizations provide input-output pairs for training data-driven surrogate models in the second stage. The surrogate models employ nonlinear dimensionality reduction using Grassmannian diffusion maps, Gaussian process regression to create a low-cost mapping between the input random vector and the reduced solution space, and geometric harmonics models for mapping between the reduced space and the Grassmann manifold. It is demonstrated that the surrogate models provide highly accurate approximations of the computational model while significantly reducing the computational cost. Monte Carlo simulations of the surrogate models are used for uncertainty propagation. UQ of strain fields highlight significant spatial variation in model uncertainty, and reveal key differences in uncertainty among commonly used strain-based brain injury predictor variables.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge