Yoshihisa Shinagawa
Fast Multi-Organ Fine Segmentation in CT Images with Hierarchical Sparse Sampling and Residual Transformer
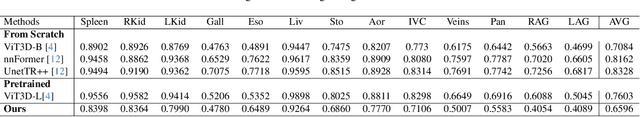
Nov 11, 2025Abstract:Multi-organ segmentation of 3D medical images is fundamental with meaningful applications in various clinical automation pipelines. Although deep learning has achieved superior performance, the time and memory consumption of segmenting the entire 3D volume voxel by voxel using neural networks can be huge. Classifiers have been developed as an alternative in cases with certain points of interest, but the trade-off between speed and accuracy remains an issue. Thus, we propose a novel fast multi-organ segmentation framework with the usage of hierarchical sparse sampling and a Residual Transformer. Compared with whole-volume analysis, the hierarchical sparse sampling strategy could successfully reduce computation time while preserving a meaningful hierarchical context utilizing multiple resolution levels. The architecture of the Residual Transformer segmentation network could extract and combine information from different levels of information in the sparse descriptor while maintaining a low computational cost. In an internal data set containing 10,253 CT images and the public dataset TotalSegmentator, the proposed method successfully improved qualitative and quantitative segmentation performance compared to the current fast organ classifier, with fast speed at the level of ~2.24 seconds on CPU hardware. The potential of achieving real-time fine organ segmentation is suggested.
BodyGPS: Anatomical Positioning System
May 12, 2025Abstract:We introduce a new type of foundational model for parsing human anatomy in medical images that works for different modalities. It supports supervised or unsupervised training and can perform matching, registration, classification, or segmentation with or without user interaction. We achieve this by training a neural network estimator that maps query locations to atlas coordinates via regression. Efficiency is improved by sparsely sampling the input, enabling response times of less than 1 ms without additional accelerator hardware. We demonstrate the utility of the algorithm in both CT and MRI modalities.
Evaluating unsupervised contrastive learning framework for MRI sequences classification
Jan 12, 2025
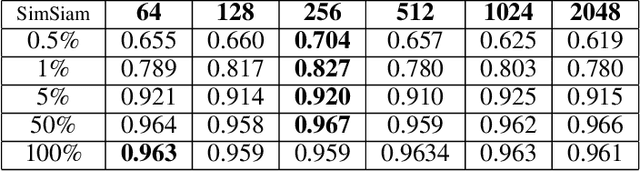
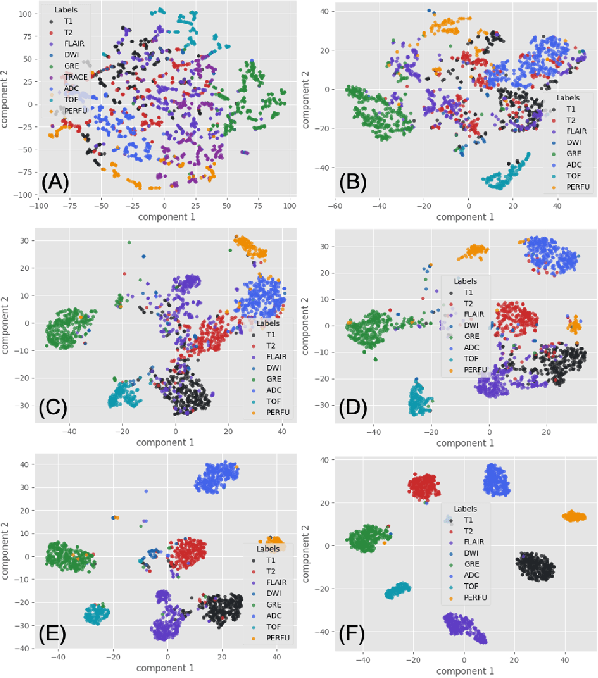
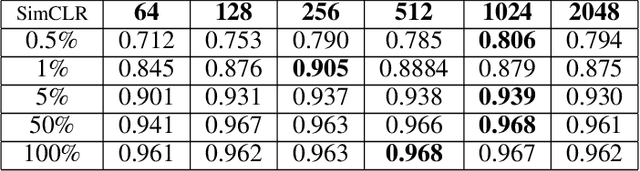
Abstract:The automatic identification of Magnetic Resonance Imaging (MRI) sequences can streamline clinical workflows by reducing the time radiologists spend manually sorting and identifying sequences, thereby enabling faster diagnosis and treatment planning for patients. However, the lack of standardization in the parameters of MRI scans poses challenges for automated systems and complicates the generation and utilization of datasets for machine learning research. To address this issue, we propose a system for MRI sequence identification using an unsupervised contrastive deep learning framework. By training a convolutional neural network based on the ResNet-18 architecture, our system classifies nine common MRI sequence types as a 9-class classification problem. The network was trained using an in-house internal dataset and validated on several public datasets, including BraTS, ADNI, Fused Radiology-Pathology Prostate Dataset, the Breast Cancer Dataset (ACRIN), among others, encompassing diverse acquisition protocols and requiring only 2D slices for training. Our system achieves a classification accuracy of over 0.95 across the nine most common MRI sequence types.
Real Time Multi Organ Classification on Computed Tomography Images
Apr 29, 2024
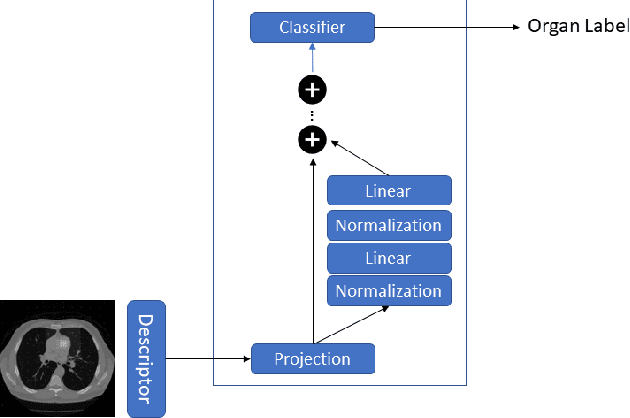

Abstract:Organ segmentation is a fundamental task in medical imaging, and it is useful for many clinical automation pipelines. Typically, the process involves segmenting the entire volume, which can be unnecessary when the points of interest are limited. In those cases, a classifier could be used instead of segmentation. However, there is an inherent trade-off between the context size and the speed of classifiers. To address this issue, we propose a new method that employs a data selection strategy with sparse sampling across a wide field of view without image resampling. This sparse sampling strategy makes it possible to classify voxels into multiple organs in real time without using accelerators. Although our method is an independent classifier, it can generate full segmentation by querying grid locations at any resolution. We have compared our method with existing segmentation techniques, demonstrating its potential for superior runtime in practical applications in medical imaging.
A Hierarchical Descriptor Framework for On-the-Fly Anatomical Location Matching between Longitudinal Studies
Aug 11, 2023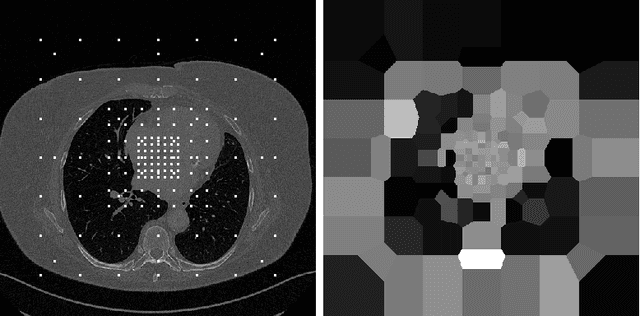
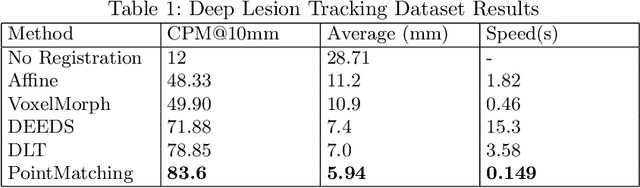
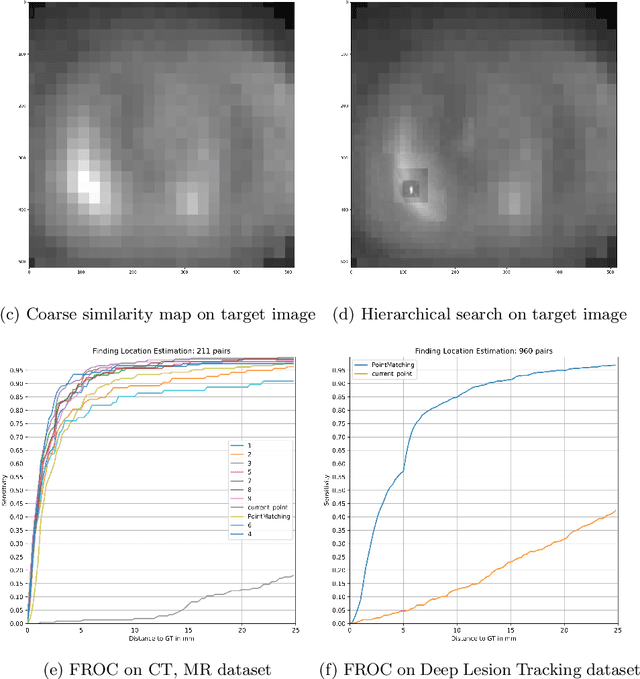
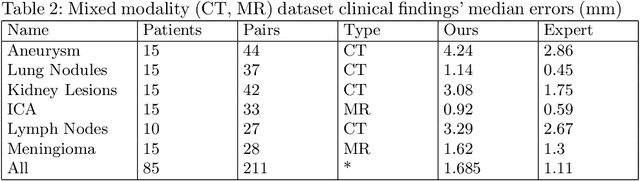
Abstract:We propose a method to match anatomical locations between pairs of medical images in longitudinal comparisons. The matching is made possible by computing a descriptor of the query point in a source image based on a hierarchical sparse sampling of image intensities that encode the location information. Then, a hierarchical search operation finds the corresponding point with the most similar descriptor in the target image. This simple yet powerful strategy reduces the computational time of mapping points to a millisecond scale on a single CPU. Thus, radiologists can compare similar anatomical locations in near real-time without requiring extra architectural costs for precomputing or storing deformation fields from registrations. Our algorithm does not require prior training, resampling, segmentation, or affine transformation steps. We have tested our algorithm on the recently published Deep Lesion Tracking dataset annotations. We observed more accurate matching compared to Deep Lesion Tracker while being 24 times faster than the most precise algorithm reported therein. We also investigated the matching accuracy on CT and MR modalities and compared the proposed algorithm's accuracy against ground truth consolidated from multiple radiologists.
Dual-Domain Cross-Iteration Squeeze-Excitation Network for Sparse Reconstruction of Brain MRI
Oct 05, 2022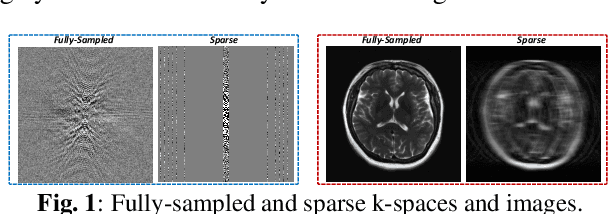
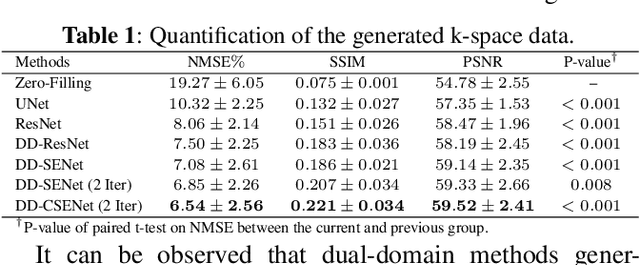
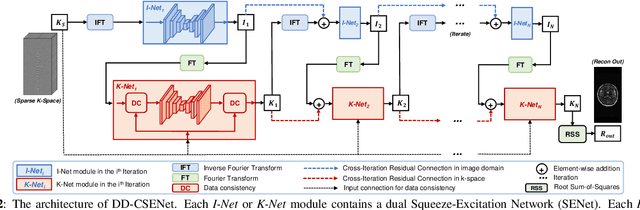
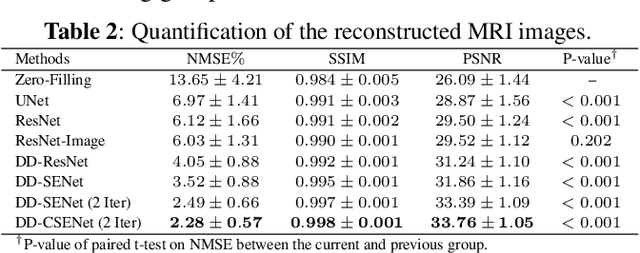
Abstract:Magnetic resonance imaging (MRI) is one of the most commonly applied tests in neurology and neurosurgery. However, the utility of MRI is largely limited by its long acquisition time, which might induce many problems including patient discomfort and motion artifacts. Acquiring fewer k-space sampling is a potential solution to reducing the total scanning time. However, it can lead to severe aliasing reconstruction artifacts and thus affect the clinical diagnosis. Nowadays, deep learning has provided new insights into the sparse reconstruction of MRI. In this paper, we present a new approach to this problem that iteratively fuses the information of k-space and MRI images using novel dual Squeeze-Excitation Networks and Cross-Iteration Residual Connections. This study included 720 clinical multi-coil brain MRI cases adopted from the open-source deidentified fastMRI Dataset. 8-folder downsampling rate was applied to generate the sparse k-space. Results showed that the average reconstruction error over 120 testing cases by our proposed method was 2.28%, which outperformed the existing image-domain prediction (6.03%, p<0.001), k-space synthesis (6.12%, p<0.001), and dual-domain feature fusion (4.05%, p<0.001).
Supervised Understanding of Word Embeddings
Jun 23, 2020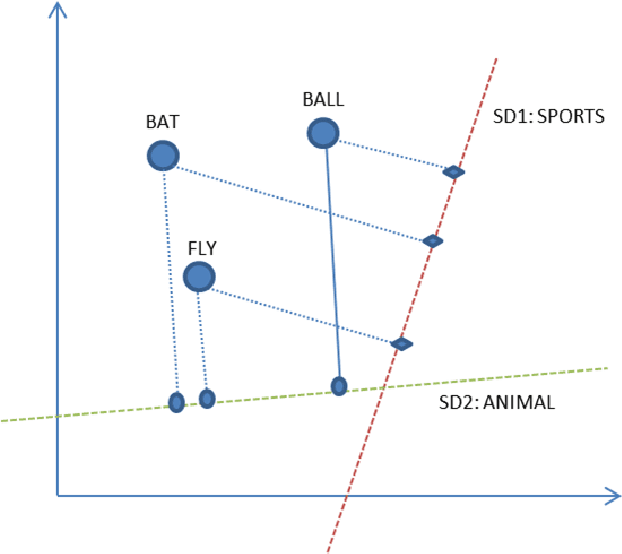

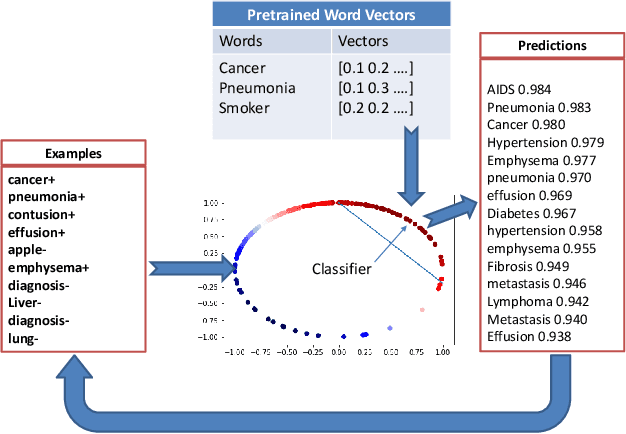

Abstract:Pre-trained word embeddings are widely used for transfer learning in natural language processing. The embeddings are continuous and distributed representations of the words that preserve their similarities in compact Euclidean spaces. However, the dimensions of these spaces do not provide any clear interpretation. In this study, we have obtained supervised projections in the form of the linear keyword-level classifiers on word embeddings. We have shown that the method creates interpretable projections of original embedding dimensions. Activations of the trained classifier nodes correspond to a subset of the words in the vocabulary. Thus, they behave similarly to the dictionary features while having the merit of continuous value output. Additionally, such dictionaries can be grown iteratively with multiple rounds by adding expert labels on top-scoring words to an initial collection of the keywords. Also, the same classifiers can be applied to aligned word embeddings in other languages to obtain corresponding dictionaries. In our experiments, we have shown that initializing higher-order networks with these classifier weights gives more accurate models for downstream NLP tasks. We further demonstrate the usefulness of supervised dimensions in revealing the polysemous nature of a keyword of interest by projecting it's embedding using learned classifiers in different sub-spaces.
Hierarchical Latent Word Clustering
Jan 20, 2016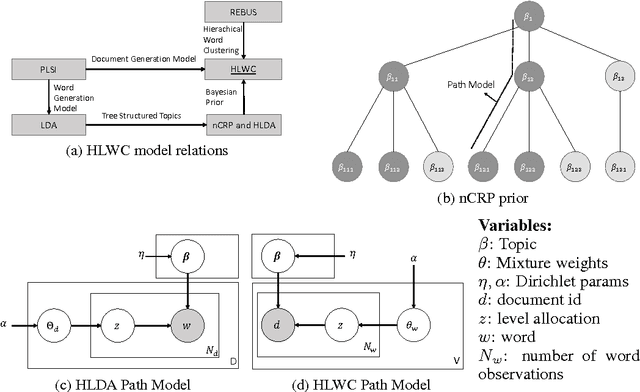
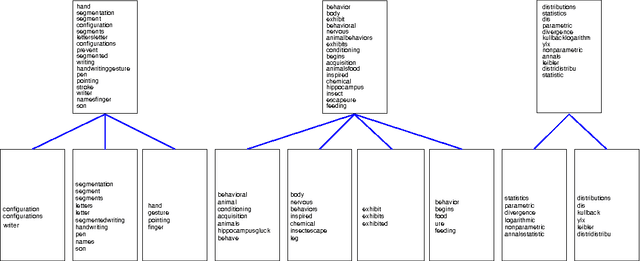
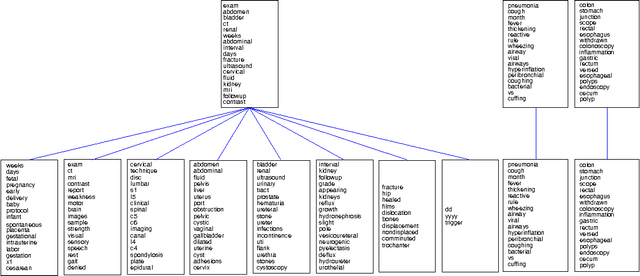
Abstract:This paper presents a new Bayesian non-parametric model by extending the usage of Hierarchical Dirichlet Allocation to extract tree structured word clusters from text data. The inference algorithm of the model collects words in a cluster if they share similar distribution over documents. In our experiments, we observed meaningful hierarchical structures on NIPS corpus and radiology reports collected from public repositories.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge