Hirotsugu Takabatake
Realistic Endoscopic Image Generation Method Using Virtual-to-real Image-domain Translation
Jan 13, 2022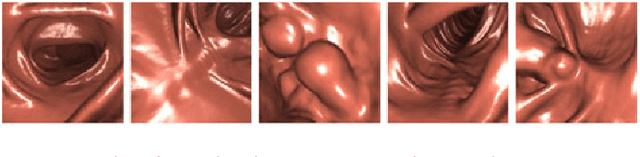

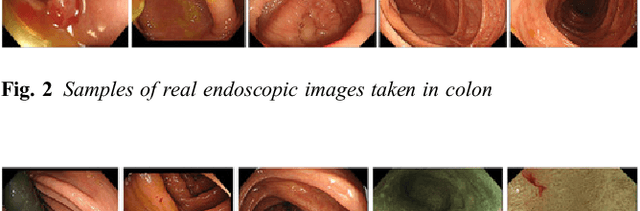
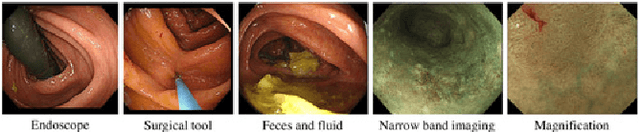
Abstract:This paper proposes a realistic image generation method for visualization in endoscopic simulation systems. Endoscopic diagnosis and treatment are performed in many hospitals. To reduce complications related to endoscope insertions, endoscopic simulation systems are used for training or rehearsal of endoscope insertions. However, current simulation systems generate non-realistic virtual endoscopic images. To improve the value of the simulation systems, improvement of reality of their generated images is necessary. We propose a realistic image generation method for endoscopic simulation systems. Virtual endoscopic images are generated by using a volume rendering method from a CT volume of a patient. We improve the reality of the virtual endoscopic images using a virtual-to-real image-domain translation technique. The image-domain translator is implemented as a fully convolutional network (FCN). We train the FCN by minimizing a cycle consistency loss function. The FCN is trained using unpaired virtual and real endoscopic images. To obtain high quality image-domain translation results, we perform an image cleansing to the real endoscopic image set. We tested to use the shallow U-Net, U-Net, deep U-Net, and U-Net having residual units as the image-domain translator. The deep U-Net and U-Net having residual units generated quite realistic images.
* Accepted paper as an oral presentation at the Joint MICCAI workshop MIAR | AE-CAI | CARE 2019
Depth Estimation from Single-shot Monocular Endoscope Image Using Image Domain Adaptation And Edge-Aware Depth Estimation
Jan 12, 2022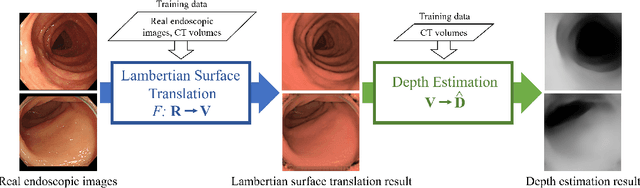

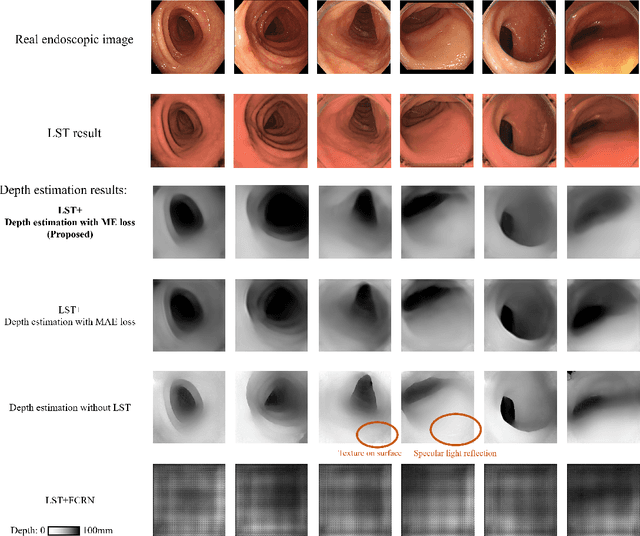

Abstract:We propose a depth estimation method from a single-shot monocular endoscopic image using Lambertian surface translation by domain adaptation and depth estimation using multi-scale edge loss. We employ a two-step estimation process including Lambertian surface translation from unpaired data and depth estimation. The texture and specular reflection on the surface of an organ reduce the accuracy of depth estimations. We apply Lambertian surface translation to an endoscopic image to remove these texture and reflections. Then, we estimate the depth by using a fully convolutional network (FCN). During the training of the FCN, improvement of the object edge similarity between an estimated image and a ground truth depth image is important for getting better results. We introduced a muti-scale edge loss function to improve the accuracy of depth estimation. We quantitatively evaluated the proposed method using real colonoscopic images. The estimated depth values were proportional to the real depth values. Furthermore, we applied the estimated depth images to automated anatomical location identification of colonoscopic images using a convolutional neural network. The identification accuracy of the network improved from 69.2% to 74.1% by using the estimated depth images.
* Accepted paper as an oral presentation at Joint MICCAI workshop 2021, AE-CAI/CARE/OR2.0
Micro CT Image-Assisted Cross Modality Super-Resolution of Clinical CT Images Utilizing Synthesized Training Dataset
Oct 20, 2020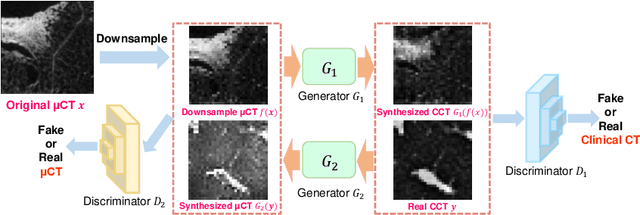
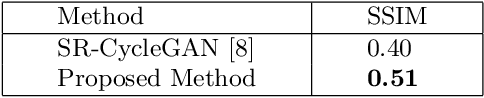
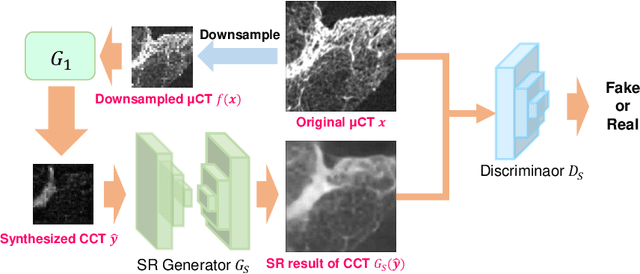

Abstract:This paper proposes a novel, unsupervised super-resolution (SR) approach for performing the SR of a clinical CT into the resolution level of a micro CT ($\mu$CT). The precise non-invasive diagnosis of lung cancer typically utilizes clinical CT data. Due to the resolution limitations of clinical CT (about $0.5 \times 0.5 \times 0.5$ mm$^3$), it is difficult to obtain enough pathological information such as the invasion area at alveoli level. On the other hand, $\mu$CT scanning allows the acquisition of volumes of lung specimens with much higher resolution ($50 \times 50 \times 50 \mu {\rm m}^3$ or higher). Thus, super-resolution of clinical CT volume may be helpful for diagnosis of lung cancer. Typical SR methods require aligned pairs of low-resolution (LR) and high-resolution (HR) images for training. Unfortunately, obtaining paired clinical CT and $\mu$CT volumes of human lung tissues is infeasible. Unsupervised SR methods are required that do not need paired LR and HR images. In this paper, we create corresponding clinical CT-$\mu$CT pairs by simulating clinical CT images from $\mu$CT images by modified CycleGAN. After this, we use simulated clinical CT-$\mu$CT image pairs to train an SR network based on SRGAN. Finally, we use the trained SR network to perform SR of the clinical CT images. We compare our proposed method with another unsupervised SR method for clinical CT images named SR-CycleGAN. Experimental results demonstrate that the proposed method can successfully perform SR of clinical CT images of lung cancer patients with $\mu$CT level resolution, and quantitatively and qualitatively outperformed conventional method (SR-CycleGAN), improving the SSIM (structure similarity) form 0.40 to 0.51.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge