John Virostko
Lifespan Pancreas Morphology for Control vs Type 2 Diabetes using AI on Largescale Clinical Imaging
Aug 20, 2025Abstract:Purpose: Understanding how the pancreas changes is critical for detecting deviations in type 2 diabetes and other pancreatic disease. We measure pancreas size and shape using morphological measurements from ages 0 to 90. Our goals are to 1) identify reliable clinical imaging modalities for AI-based pancreas measurement, 2) establish normative morphological aging trends, and 3) detect potential deviations in type 2 diabetes. Approach: We analyzed a clinically acquired dataset of 2533 patients imaged with abdominal CT or MRI. We resampled the scans to 3mm isotropic resolution, segmented the pancreas using automated methods, and extracted 13 morphological pancreas features across the lifespan. First, we assessed CT and MRI measurements to determine which modalities provide consistent lifespan trends. Second, we characterized distributions of normative morphological patterns stratified by age group and sex. Third, we used GAMLSS regression to model pancreas morphology trends in 1350 patients matched for age, sex, and type 2 diabetes status to identify any deviations from normative aging associated with type 2 diabetes. Results: When adjusting for confounders, the aging trends for 10 of 13 morphological features were significantly different between patients with type 2 diabetes and non-diabetic controls (p < 0.05 after multiple comparisons corrections). Additionally, MRI appeared to yield different pancreas measurements than CT using our AI-based method. Conclusions: We provide lifespan trends demonstrating that the size and shape of the pancreas is altered in type 2 diabetes using 675 control patients and 675 diabetes patients. Moreover, our findings reinforce that the pancreas is smaller in type 2 diabetes. Additionally, we contribute a reference of lifespan pancreas morphology from a large cohort of non-diabetic control patients in a clinical setting.
Influence of Early through Late Fusion on Pancreas Segmentation from Imperfectly Registered Multimodal MRI
Sep 06, 2024Abstract:Multimodal fusion promises better pancreas segmentation. However, where to perform fusion in models is still an open question. It is unclear if there is a best location to fuse information when analyzing pairs of imperfectly aligned images. Two main alignment challenges in this pancreas segmentation study are 1) the pancreas is deformable and 2) breathing deforms the abdomen. Even after image registration, relevant deformations are often not corrected. We examine how early through late fusion impacts pancreas segmentation. We used 353 pairs of T2-weighted (T2w) and T1-weighted (T1w) abdominal MR images from 163 subjects with accompanying pancreas labels. We used image registration (deeds) to align the image pairs. We trained a collection of basic UNets with different fusion points, spanning from early to late, to assess how early through late fusion influenced segmentation performance on imperfectly aligned images. We assessed generalization of fusion points on nnUNet. The single-modality T2w baseline using a basic UNet model had a Dice score of 0.73, while the same baseline on the nnUNet model achieved 0.80. For the basic UNet, the best fusion approach occurred in the middle of the encoder (early/mid fusion), which led to a statistically significant improvement of 0.0125 on Dice score compared to the baseline. For the nnUNet, the best fusion approach was na\"ive image concatenation before the model (early fusion), which resulted in a statistically significant Dice score increase of 0.0021 compared to baseline. Fusion in specific blocks can improve performance, but the best blocks for fusion are model specific, and the gains are small. In imperfectly registered datasets, fusion is a nuanced problem, with the art of design remaining vital for uncovering potential insights. Future innovation is needed to better address fusion in cases of imperfect alignment of abdominal image pairs.
An untrained deep learning method for reconstructing dynamic magnetic resonance images from accelerated model-based data
May 04, 2022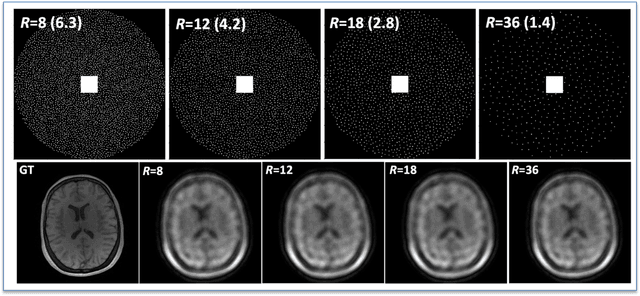

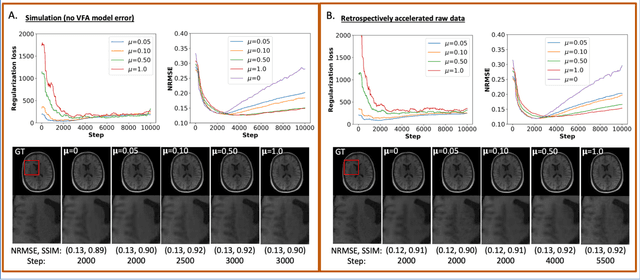

Abstract:The purpose of this work is to implement physics-based regularization as a stopping condition in tuning an untrained deep neural network for reconstructing MR images from accelerated data. The ConvDecoder neural network was trained with a physics-based regularization term incorporating the spoiled gradient echo equation that describes variable-flip angle (VFA) data. Fully-sampled VFA k-space data were retrospectively accelerated by factors of R={8,12,18,36} and reconstructed with ConvDecoder (CD), ConvDecoder with the proposed regularization (CD+r), locally low-rank (LR) reconstruction, and compressed sensing with L1-wavelet regularization (L1). Final images from CD+r training were evaluated at the \emph{argmin} of the regularization loss; whereas the CD, LR, and L1 reconstructions were chosen optimally based on ground truth data. The performance measures used were the normalized root-mean square error, the concordance correlation coefficient (CCC), and the structural similarity index (SSIM). The CD+r reconstructions, chosen using the stopping condition, yielded SSIMs that were similar to the CD (p=0.47) and LR SSIMs (p=0.95) across R and that were significantly higher than the L1 SSIMs (p=0.04). The CCC values for the CD+r T1 maps across all R and subjects were greater than those corresponding to the L1 (p=0.15) and LR (p=0.13) T1 maps, respectively. For R > 12 (<4.2 minutes scan time), L1 and LR T1 maps exhibit a loss of spatially refined details compared to CD+r. We conclude that the use of an untrained neural network together with a physics-based regularization loss shows promise as a measure for determining the optimal stopping point in training without relying on fully-sampled ground truth data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge