Ming-Kai Chen
Anatomically and Metabolically Informed Diffusion for Unified Denoising and Segmentation in Low-Count PET Imaging
Mar 17, 2025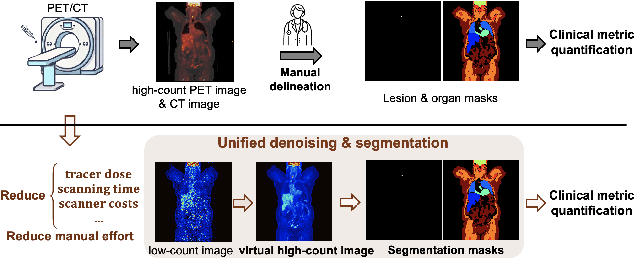
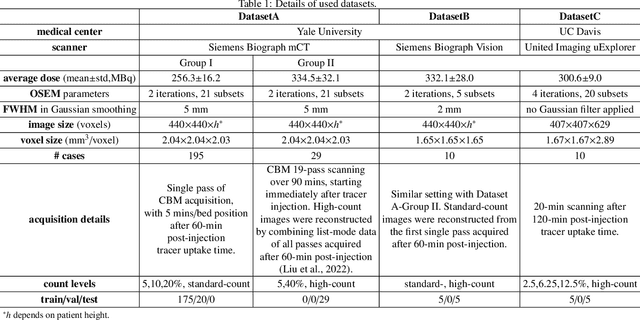
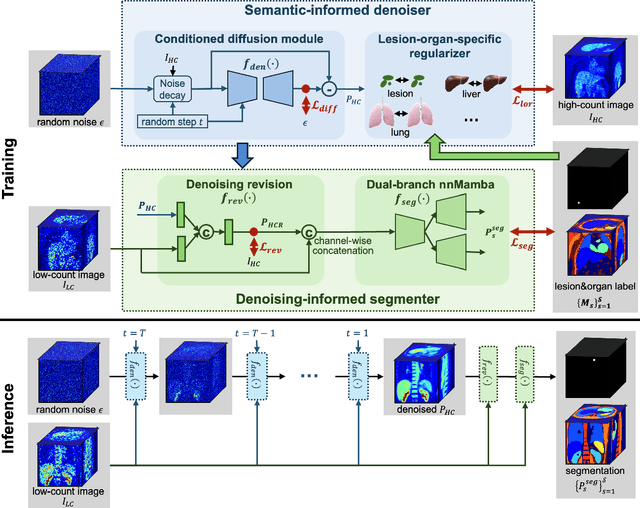
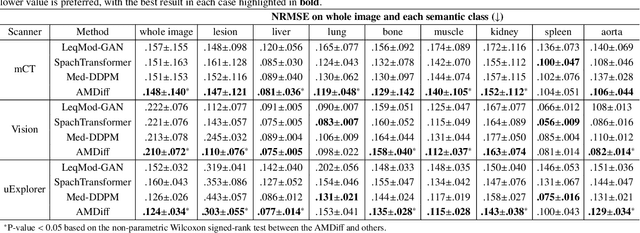
Abstract:Positron emission tomography (PET) image denoising, along with lesion and organ segmentation, are critical steps in PET-aided diagnosis. However, existing methods typically treat these tasks independently, overlooking inherent synergies between them as correlated steps in the analysis pipeline. In this work, we present the anatomically and metabolically informed diffusion (AMDiff) model, a unified framework for denoising and lesion/organ segmentation in low-count PET imaging. By integrating multi-task functionality and exploiting the mutual benefits of these tasks, AMDiff enables direct quantification of clinical metrics, such as total lesion glycolysis (TLG), from low-count inputs. The AMDiff model incorporates a semantic-informed denoiser based on diffusion strategy and a denoising-informed segmenter utilizing nnMamba architecture. The segmenter constrains denoised outputs via a lesion-organ-specific regularizer, while the denoiser enhances the segmenter by providing enriched image information through a denoising revision module. These components are connected via a warming-up mechanism to optimize multitask interactions. Experiments on multi-vendor, multi-center, and multi-noise-level datasets demonstrate the superior performance of AMDiff. For test cases below 20% of the clinical count levels from participating sites, AMDiff achieves TLG quantification biases of -26.98%, outperforming its ablated versions which yield biases of -35.85% (without the lesion-organ-specific regularizer) and -40.79% (without the denoising revision module).
Synthesizing Multi-Tracer PET Images for Alzheimer's Disease Patients using a 3D Unified Anatomy-aware Cyclic Adversarial Network
Jul 12, 2021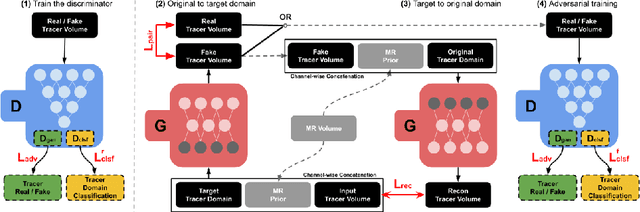
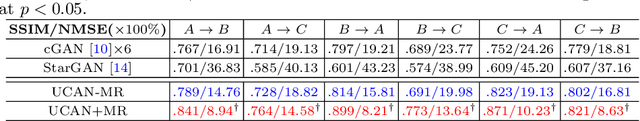
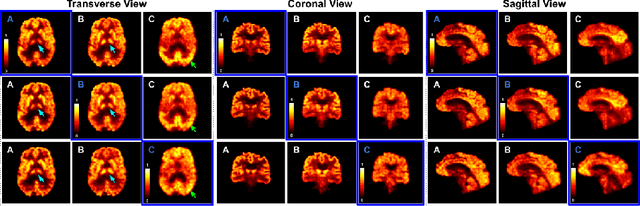
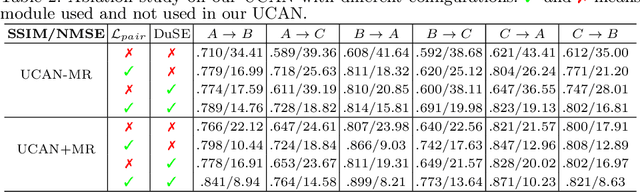
Abstract:Positron Emission Tomography (PET) is an important tool for studying Alzheimer's disease (AD). PET scans can be used as diagnostics tools, and to provide molecular characterization of patients with cognitive disorders. However, multiple tracers are needed to measure glucose metabolism (18F-FDG), synaptic vesicle protein (11C-UCB-J), and $\beta$-amyloid (11C-PiB). Administering multiple tracers to patient will lead to high radiation dose and cost. In addition, access to PET scans using new or less-available tracers with sophisticated production methods and short half-life isotopes may be very limited. Thus, it is desirable to develop an efficient multi-tracer PET synthesis model that can generate multi-tracer PET from single-tracer PET. Previous works on medical image synthesis focus on one-to-one fixed domain translations, and cannot simultaneously learn the feature from multi-tracer domains. Given 3 or more tracers, relying on previous methods will also create a heavy burden on the number of models to be trained. To tackle these issues, we propose a 3D unified anatomy-aware cyclic adversarial network (UCAN) for translating multi-tracer PET volumes with one unified generative model, where MR with anatomical information is incorporated. Evaluations on a multi-tracer PET dataset demonstrate the feasibility that our UCAN can generate high-quality multi-tracer PET volumes, with NMSE less than 15% for all PET tracers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge