Jing Jiao
TinyUSFM: Towards Compact and Efficient Ultrasound Foundation Models
Oct 22, 2025Abstract:Foundation models for medical imaging demonstrate superior generalization capabilities across diverse anatomical structures and clinical applications. Their outstanding performance relies on substantial computational resources, limiting deployment in resource-constrained clinical environments. This paper presents TinyUSFM, the first lightweight ultrasound foundation model that maintains superior organ versatility and task adaptability of our large-scale Ultrasound Foundation Model (USFM) through knowledge distillation with strategically curated small datasets, delivering significant computational efficiency without sacrificing performance. Considering the limited capacity and representation ability of lightweight models, we propose a feature-gradient driven coreset selection strategy to curate high-quality compact training data, avoiding training degradation from low-quality redundant images. To preserve the essential spatial and frequency domain characteristics during knowledge transfer, we develop domain-separated masked image modeling assisted consistency-driven dynamic distillation. This novel framework adaptively transfers knowledge from large foundation models by leveraging teacher model consistency across different domain masks, specifically tailored for ultrasound interpretation. For evaluation, we establish the UniUS-Bench, the largest publicly available ultrasound benchmark comprising 8 classification and 10 segmentation datasets across 15 organs. Using only 200K images in distillation, TinyUSFM matches USFM's performance with just 6.36% of parameters and 6.40% of GFLOPs. TinyUSFM significantly outperforms the vanilla model by 9.45% in classification and 7.72% in segmentation, surpassing all state-of-the-art lightweight models, and achieving 84.91% average classification accuracy and 85.78% average segmentation Dice score across diverse medical devices and centers.
Chest-Diffusion: A Light-Weight Text-to-Image Model for Report-to-CXR Generation
Jun 30, 2024Abstract:Text-to-image generation has important implications for generation of diverse and controllable images. Several attempts have been made to adapt Stable Diffusion (SD) to the medical domain. However, the large distribution difference between medical reports and natural texts, as well as high computational complexity in common stable diffusion limit the authenticity and feasibility of the generated medical images. To solve above problems, we propose a novel light-weight transformer-based diffusion model learning framework, Chest-Diffusion, for report-to-CXR generation. Chest-Diffusion employs a domain-specific text encoder to obtain accurate and expressive text features to guide image generation, improving the authenticity of the generated images. Meanwhile, we introduce a light-weight transformer architecture as the denoising model, reducing the computational complexity of the diffusion model. Experiments demonstrate that our Chest-Diffusion achieves the lowest FID score 24.456, under the computation budget of 118.918 GFLOPs, which is nearly one-third of the computational complexity of SD.
USFM: A Universal Ultrasound Foundation Model Generalized to Tasks and Organs towards Label Efficient Image Analysis
Jan 02, 2024Abstract:Inadequate generality across different organs and tasks constrains the application of ultrasound (US) image analysis methods in smart healthcare. Building a universal US foundation model holds the potential to address these issues. Nevertheless, the development of such foundational models encounters intrinsic challenges in US analysis, i.e., insufficient databases, low quality, and ineffective features. In this paper, we present a universal US foundation model, named USFM, generalized to diverse tasks and organs towards label efficient US image analysis. First, a large-scale Multi-organ, Multi-center, and Multi-device US database was built, comprehensively containing over two million US images. Organ-balanced sampling was employed for unbiased learning. Then, USFM is self-supervised pre-trained on the sufficient US database. To extract the effective features from low-quality US images, we proposed a spatial-frequency dual masked image modeling method. A productive spatial noise addition-recovery approach was designed to learn meaningful US information robustly, while a novel frequency band-stop masking learning approach was also employed to extract complex, implicit grayscale distribution and textural variations. Extensive experiments were conducted on the various tasks of segmentation, classification, and image enhancement from diverse organs and diseases. Comparisons with representative US image analysis models illustrate the universality and effectiveness of USFM. The label efficiency experiments suggest the USFM obtains robust performance with only 20% annotation, laying the groundwork for the rapid development of US models in clinical practices.
FetReg2021: A Challenge on Placental Vessel Segmentation and Registration in Fetoscopy
Jun 30, 2022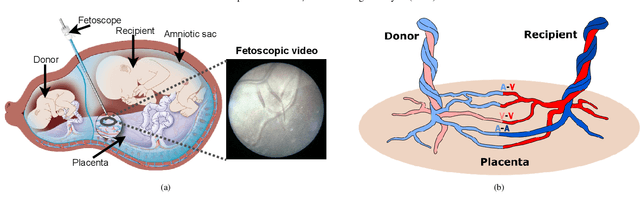
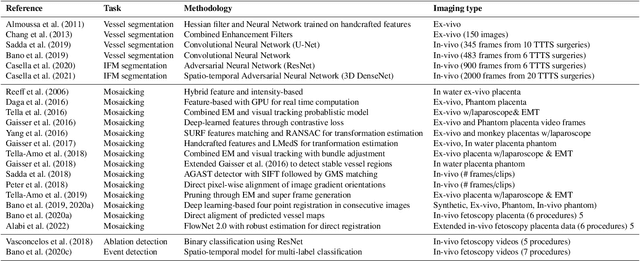
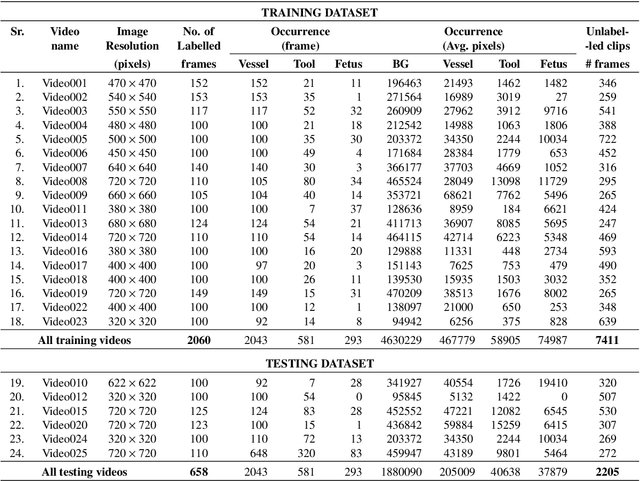
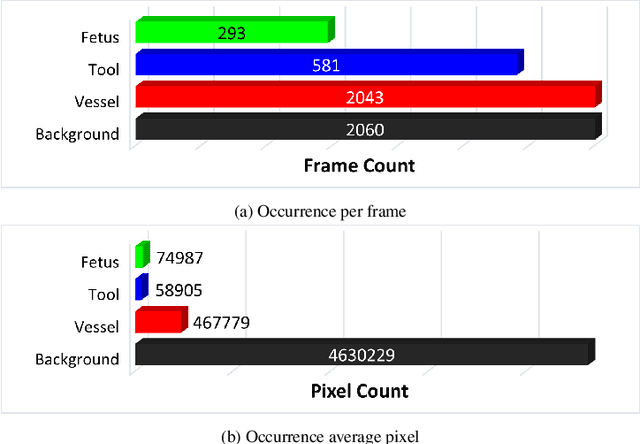
Abstract:Fetoscopy laser photocoagulation is a widely adopted procedure for treating Twin-to-Twin Transfusion Syndrome (TTTS). The procedure involves photocoagulation pathological anastomoses to regulate blood exchange among twins. The procedure is particularly challenging due to the limited field of view, poor manoeuvrability of the fetoscope, poor visibility, and variability in illumination. These challenges may lead to increased surgery time and incomplete ablation. Computer-assisted intervention (CAI) can provide surgeons with decision support and context awareness by identifying key structures in the scene and expanding the fetoscopic field of view through video mosaicking. Research in this domain has been hampered by the lack of high-quality data to design, develop and test CAI algorithms. Through the Fetoscopic Placental Vessel Segmentation and Registration (FetReg2021) challenge, which was organized as part of the MICCAI2021 Endoscopic Vision challenge, we released the first largescale multicentre TTTS dataset for the development of generalized and robust semantic segmentation and video mosaicking algorithms. For this challenge, we released a dataset of 2060 images, pixel-annotated for vessels, tool, fetus and background classes, from 18 in-vivo TTTS fetoscopy procedures and 18 short video clips. Seven teams participated in this challenge and their model performance was assessed on an unseen test dataset of 658 pixel-annotated images from 6 fetoscopic procedures and 6 short clips. The challenge provided an opportunity for creating generalized solutions for fetoscopic scene understanding and mosaicking. In this paper, we present the findings of the FetReg2021 challenge alongside reporting a detailed literature review for CAI in TTTS fetoscopy. Through this challenge, its analysis and the release of multi-centre fetoscopic data, we provide a benchmark for future research in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge