Gaia Romana De Paolis
Fast Medical Shape Reconstruction via Meta-learned Implicit Neural Representations
Sep 11, 2024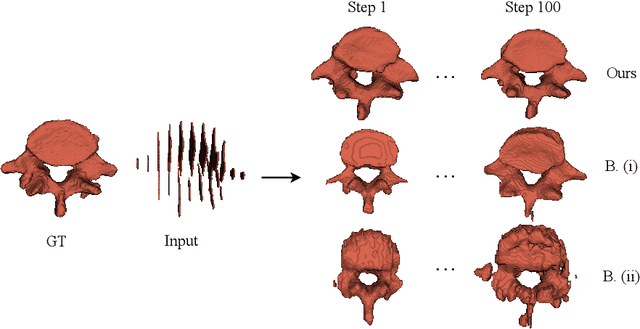
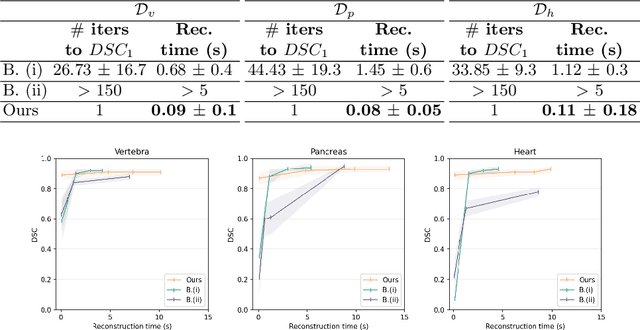
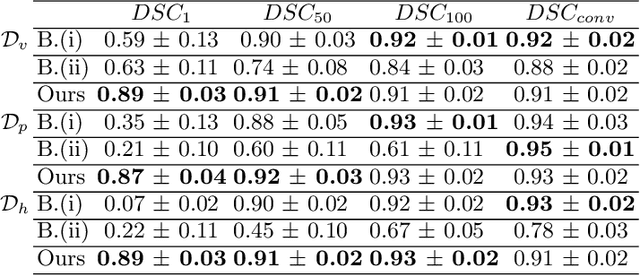

Abstract:Efficient and fast reconstruction of anatomical structures plays a crucial role in clinical practice. Minimizing retrieval and processing times not only potentially enhances swift response and decision-making in critical scenarios but also supports interactive surgical planning and navigation. Recent methods attempt to solve the medical shape reconstruction problem by utilizing implicit neural functions. However, their performance suffers in terms of generalization and computation time, a critical metric for real-time applications. To address these challenges, we propose to leverage meta-learning to improve the network parameters initialization, reducing inference time by an order of magnitude while maintaining high accuracy. We evaluate our approach on three public datasets covering different anatomical shapes and modalities, namely CT and MRI. Our experimental results show that our model can handle various input configurations, such as sparse slices with different orientations and spacings. Additionally, we demonstrate that our method exhibits strong transferable capabilities in generalizing to shape domains unobserved at training time.
PARMESAN: Parameter-Free Memory Search and Transduction for Dense Prediction Tasks
Mar 18, 2024Abstract:In this work we address flexibility in deep learning by means of transductive reasoning. For adaptation to new tasks or new data, existing methods typically involve tuning of learnable parameters or even complete re-training from scratch, rendering such approaches unflexible in practice. We argue that the notion of separating computation from memory by the means of transduction can act as a stepping stone for solving these issues. We therefore propose PARMESAN (parameter-free memory search and transduction), a scalable transduction method which leverages a memory module for solving dense prediction tasks. At inference, hidden representations in memory are being searched to find corresponding examples. In contrast to other methods, PARMESAN learns without the requirement for any continuous training or fine-tuning of learnable parameters simply by modifying the memory content. Our method is compatible with commonly used neural architectures and canonically transfers to 1D, 2D, and 3D grid-based data. We demonstrate the capabilities of our approach at complex tasks such as continual and few-shot learning. PARMESAN learns up to 370 times faster than common baselines while being on par in terms of predictive performance, knowledge retention, and data-efficiency.
Multi-scale attention-based instance segmentation for measuring crystals with large size variation
Jan 08, 2024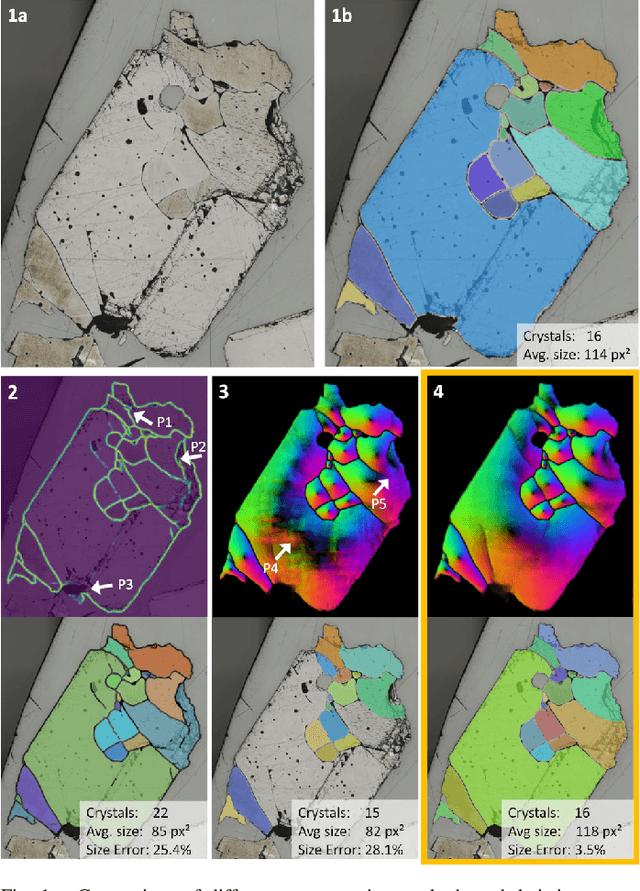
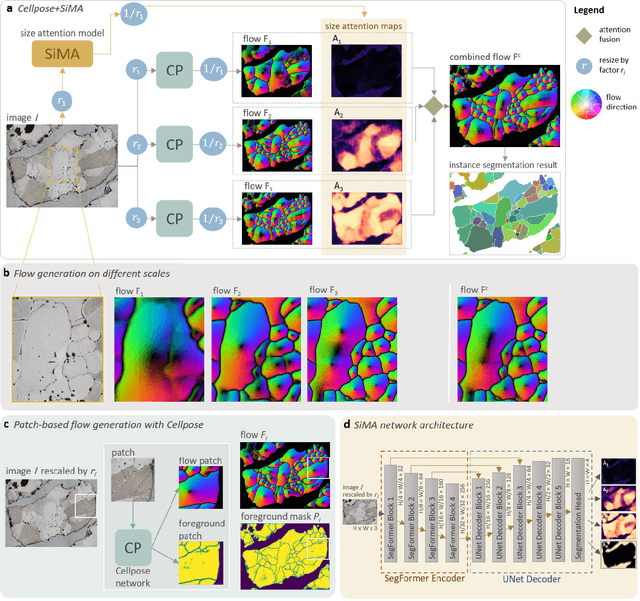
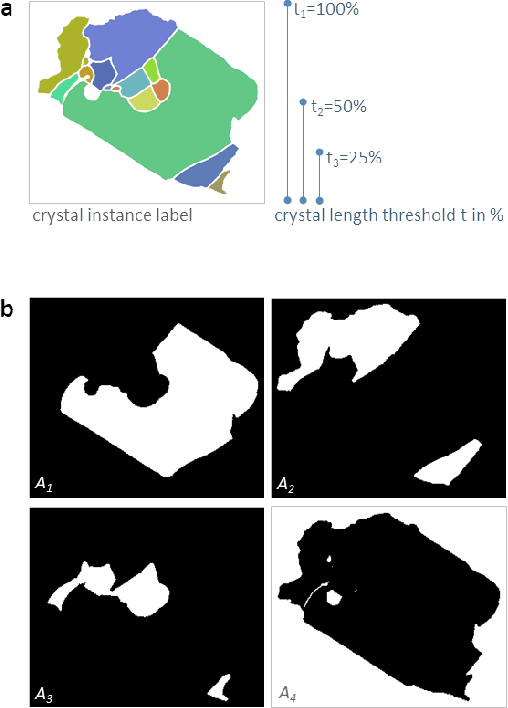
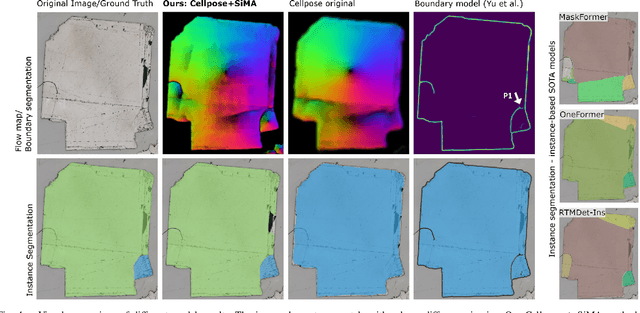
Abstract:Quantitative measurement of crystals in high-resolution images allows for important insights into underlying material characteristics. Deep learning has shown great progress in vision-based automatic crystal size measurement, but current instance segmentation methods reach their limits with images that have large variation in crystal size or hard to detect crystal boundaries. Even small image segmentation errors, such as incorrectly fused or separated segments, can significantly lower the accuracy of the measured results. Instead of improving the existing pixel-wise boundary segmentation methods, we propose to use an instance-based segmentation method, which gives more robust segmentation results to improve measurement accuracy. Our novel method enhances flow maps with a size-aware multi-scale attention module. The attention module adaptively fuses information from multiple scales and focuses on the most relevant scale for each segmented image area. We demonstrate that our proposed attention fusion strategy outperforms state-of-the-art instance and boundary segmentation methods, as well as simple average fusion of multi-scale predictions. We evaluate our method on a refractory raw material dataset of high-resolution images with large variation in crystal size and show that our model can be used to calculate the crystal size more accurately than existing methods.
FetReg2021: A Challenge on Placental Vessel Segmentation and Registration in Fetoscopy
Jun 30, 2022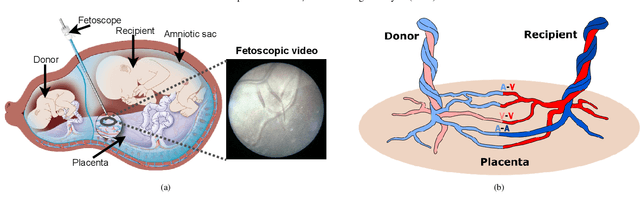
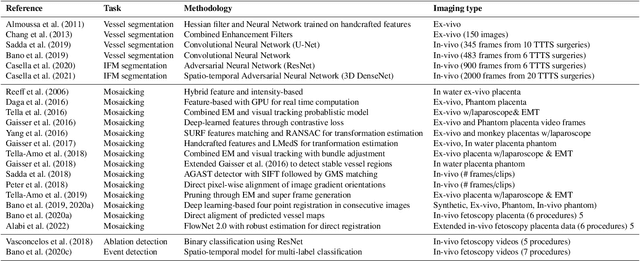
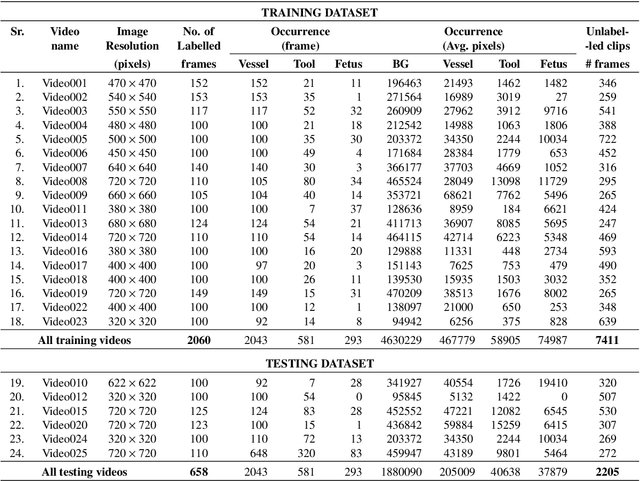
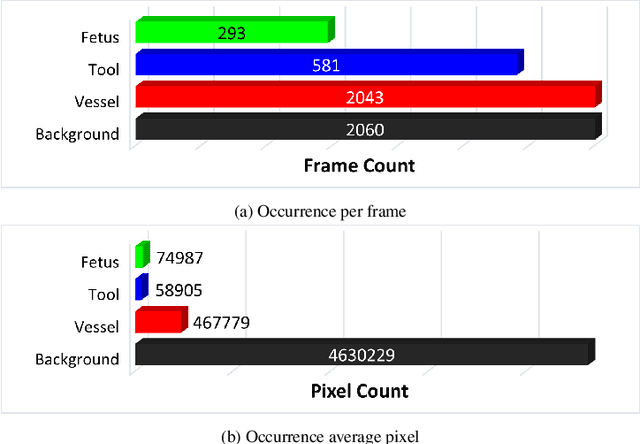
Abstract:Fetoscopy laser photocoagulation is a widely adopted procedure for treating Twin-to-Twin Transfusion Syndrome (TTTS). The procedure involves photocoagulation pathological anastomoses to regulate blood exchange among twins. The procedure is particularly challenging due to the limited field of view, poor manoeuvrability of the fetoscope, poor visibility, and variability in illumination. These challenges may lead to increased surgery time and incomplete ablation. Computer-assisted intervention (CAI) can provide surgeons with decision support and context awareness by identifying key structures in the scene and expanding the fetoscopic field of view through video mosaicking. Research in this domain has been hampered by the lack of high-quality data to design, develop and test CAI algorithms. Through the Fetoscopic Placental Vessel Segmentation and Registration (FetReg2021) challenge, which was organized as part of the MICCAI2021 Endoscopic Vision challenge, we released the first largescale multicentre TTTS dataset for the development of generalized and robust semantic segmentation and video mosaicking algorithms. For this challenge, we released a dataset of 2060 images, pixel-annotated for vessels, tool, fetus and background classes, from 18 in-vivo TTTS fetoscopy procedures and 18 short video clips. Seven teams participated in this challenge and their model performance was assessed on an unseen test dataset of 658 pixel-annotated images from 6 fetoscopic procedures and 6 short clips. The challenge provided an opportunity for creating generalized solutions for fetoscopic scene understanding and mosaicking. In this paper, we present the findings of the FetReg2021 challenge alongside reporting a detailed literature review for CAI in TTTS fetoscopy. Through this challenge, its analysis and the release of multi-centre fetoscopic data, we provide a benchmark for future research in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge