Theresa Neubauer
Fast Medical Shape Reconstruction via Meta-learned Implicit Neural Representations
Sep 11, 2024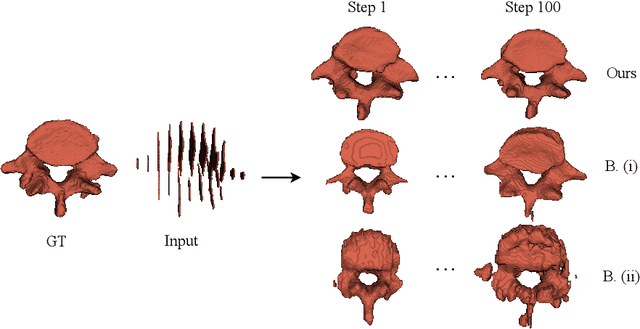
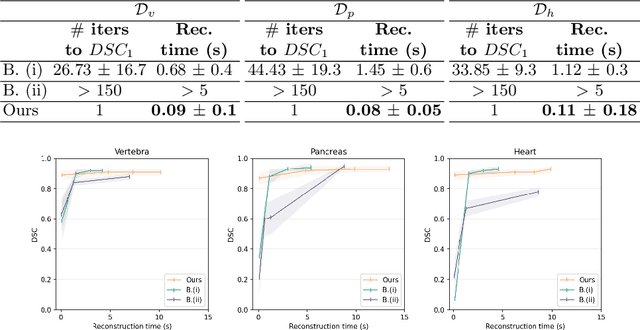
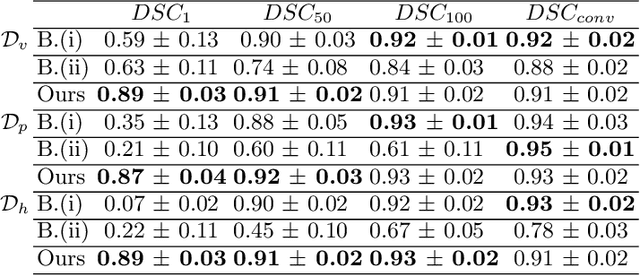

Abstract:Efficient and fast reconstruction of anatomical structures plays a crucial role in clinical practice. Minimizing retrieval and processing times not only potentially enhances swift response and decision-making in critical scenarios but also supports interactive surgical planning and navigation. Recent methods attempt to solve the medical shape reconstruction problem by utilizing implicit neural functions. However, their performance suffers in terms of generalization and computation time, a critical metric for real-time applications. To address these challenges, we propose to leverage meta-learning to improve the network parameters initialization, reducing inference time by an order of magnitude while maintaining high accuracy. We evaluate our approach on three public datasets covering different anatomical shapes and modalities, namely CT and MRI. Our experimental results show that our model can handle various input configurations, such as sparse slices with different orientations and spacings. Additionally, we demonstrate that our method exhibits strong transferable capabilities in generalizing to shape domains unobserved at training time.
PARMESAN: Parameter-Free Memory Search and Transduction for Dense Prediction Tasks
Mar 18, 2024Abstract:In this work we address flexibility in deep learning by means of transductive reasoning. For adaptation to new tasks or new data, existing methods typically involve tuning of learnable parameters or even complete re-training from scratch, rendering such approaches unflexible in practice. We argue that the notion of separating computation from memory by the means of transduction can act as a stepping stone for solving these issues. We therefore propose PARMESAN (parameter-free memory search and transduction), a scalable transduction method which leverages a memory module for solving dense prediction tasks. At inference, hidden representations in memory are being searched to find corresponding examples. In contrast to other methods, PARMESAN learns without the requirement for any continuous training or fine-tuning of learnable parameters simply by modifying the memory content. Our method is compatible with commonly used neural architectures and canonically transfers to 1D, 2D, and 3D grid-based data. We demonstrate the capabilities of our approach at complex tasks such as continual and few-shot learning. PARMESAN learns up to 370 times faster than common baselines while being on par in terms of predictive performance, knowledge retention, and data-efficiency.
Multi-scale attention-based instance segmentation for measuring crystals with large size variation
Jan 08, 2024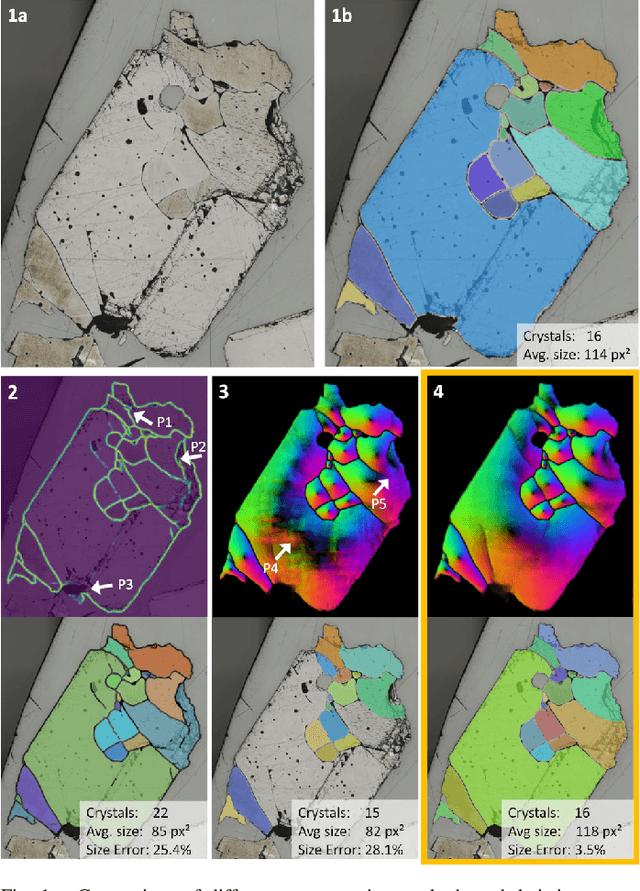
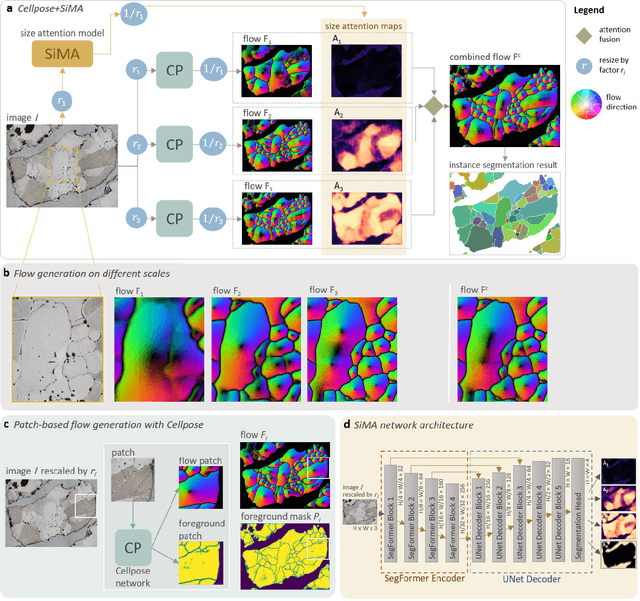
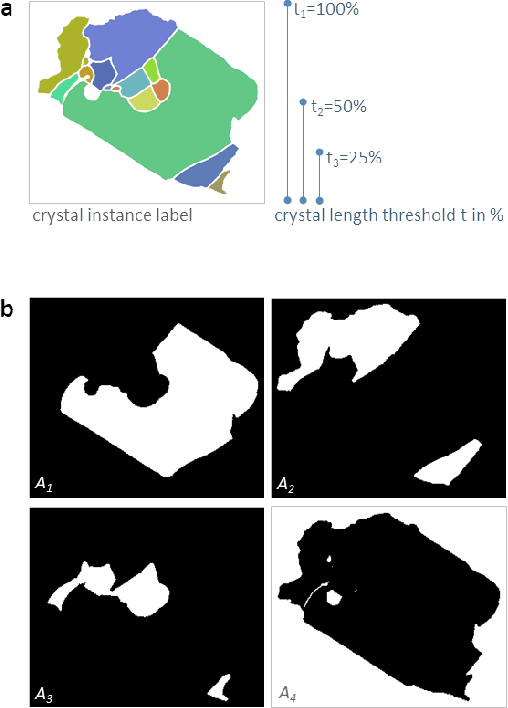
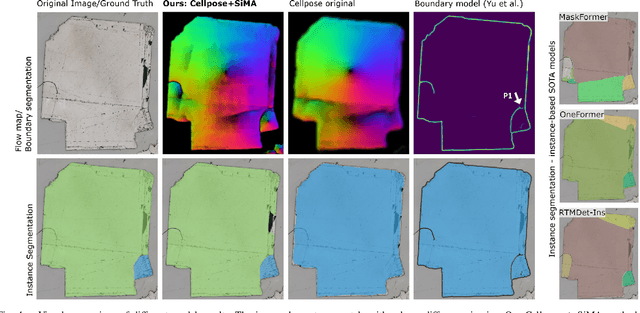
Abstract:Quantitative measurement of crystals in high-resolution images allows for important insights into underlying material characteristics. Deep learning has shown great progress in vision-based automatic crystal size measurement, but current instance segmentation methods reach their limits with images that have large variation in crystal size or hard to detect crystal boundaries. Even small image segmentation errors, such as incorrectly fused or separated segments, can significantly lower the accuracy of the measured results. Instead of improving the existing pixel-wise boundary segmentation methods, we propose to use an instance-based segmentation method, which gives more robust segmentation results to improve measurement accuracy. Our novel method enhances flow maps with a size-aware multi-scale attention module. The attention module adaptively fuses information from multiple scales and focuses on the most relevant scale for each segmented image area. We demonstrate that our proposed attention fusion strategy outperforms state-of-the-art instance and boundary segmentation methods, as well as simple average fusion of multi-scale predictions. We evaluate our method on a refractory raw material dataset of high-resolution images with large variation in crystal size and show that our model can be used to calculate the crystal size more accurately than existing methods.
Employing similarity to highlight differences: On the impact of anatomical assumptions in chest X-ray registration methods
Jan 24, 2023



Abstract:To facilitate both the detection and the interpretation of findings in chest X-rays, comparison with a previous image of the same patient is very valuable to radiologists. Today, the most common approach for deep learning methods to automatically inspect chest X-rays disregards the patient history and classifies only single images as normal or abnormal. Nevertheless, several methods for assisting in the task of comparison through image registration have been proposed in the past. However, as we illustrate, they tend to miss specific types of pathological changes like cardiomegaly and effusion. Due to assumptions on fixed anatomical structures or their measurements of registration quality, they produce unnaturally deformed warp fields impacting visualization of differences between moving and fixed images. We aim to overcome these limitations, through a new paradigm based on individual rib pair segmentation for anatomy penalized registration. Our method proves to be a natural way to limit the folding percentage of the warp field to 1/6 of the state of the art while increasing the overlap of ribs by more than 25%, implying difference images showing pathological changes overlooked by other methods. We develop an anatomically penalized convolutional multi-stage solution on the National Institutes of Health (NIH) data set, starting from less than 25 fully and 50 partly labeled training images, employing sequential instance memory segmentation with hole dropout, weak labeling, coarse-to-fine refinement and Gaussian mixture model histogram matching. We statistically evaluate the benefits of our method and highlight the limits of currently used metrics for registration of chest X-rays.
Anomaly Detection using Generative Models and Sum-Product Networks in Mammography Scans
Oct 12, 2022Abstract:Unsupervised anomaly detection models which are trained solely by healthy data, have gained importance in the recent years, as the annotation of medical data is a tedious task. Autoencoders and generative adversarial networks are the standard anomaly detection methods that are utilized to learn the data distribution. However, they fall short when it comes to inference and evaluation of the likelihood of test samples. We propose a novel combination of generative models and a probabilistic graphical model. After encoding image samples by autoencoders, the distribution of data is modeled by Random and Tensorized Sum-Product Networks ensuring exact and efficient inference at test time. We evaluate different autoencoder architectures in combination with Random and Tensorized Sum-Product Networks on mammography images using patch-wise processing and observe superior performance over utilizing the models standalone and state-of-the-art in anomaly detection for medical data.
* Submitted to DGM4MICCAI 2022 Workshop. This preprint has not undergone peer review (when applicable) or any post-submission improvements or corrections. The Version of Record of this contribution is published in LNCS 13609, and is available online at https://doi.org/10.1007/978-3-031-18576-2_8
Multi-task fusion for improving mammography screening data classification
Dec 01, 2021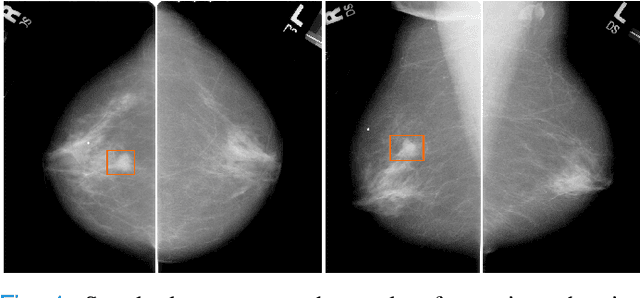
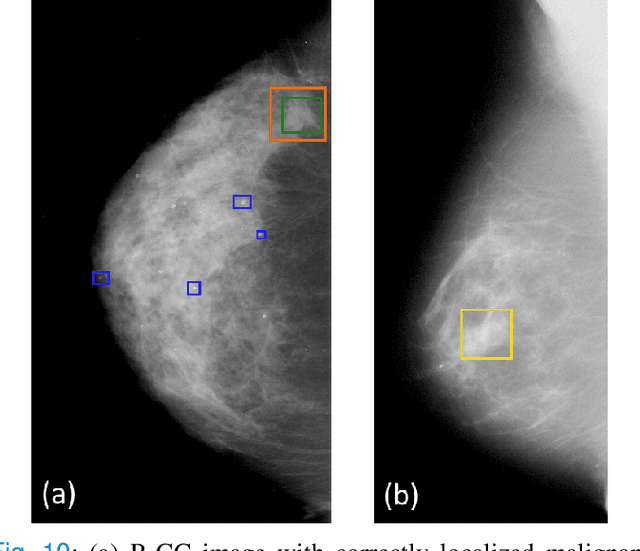
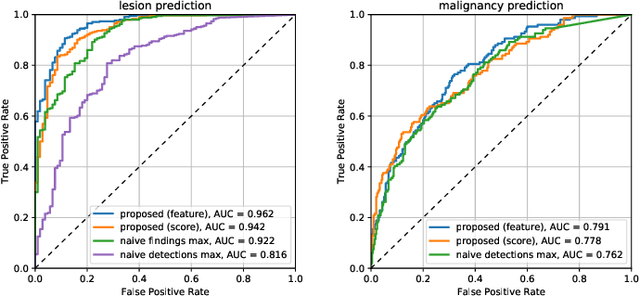
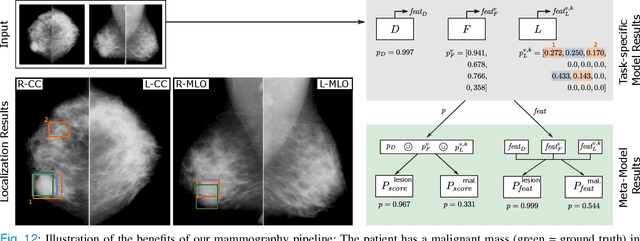
Abstract:Machine learning and deep learning methods have become essential for computer-assisted prediction in medicine, with a growing number of applications also in the field of mammography. Typically these algorithms are trained for a specific task, e.g., the classification of lesions or the prediction of a mammogram's pathology status. To obtain a comprehensive view of a patient, models which were all trained for the same task(s) are subsequently ensembled or combined. In this work, we propose a pipeline approach, where we first train a set of individual, task-specific models and subsequently investigate the fusion thereof, which is in contrast to the standard model ensembling strategy. We fuse model predictions and high-level features from deep learning models with hybrid patient models to build stronger predictors on patient level. To this end, we propose a multi-branch deep learning model which efficiently fuses features across different tasks and mammograms to obtain a comprehensive patient-level prediction. We train and evaluate our full pipeline on public mammography data, i.e., DDSM and its curated version CBIS-DDSM, and report an AUC score of 0.962 for predicting the presence of any lesion and 0.791 for predicting the presence of malignant lesions on patient level. Overall, our fusion approaches improve AUC scores significantly by up to 0.04 compared to standard model ensembling. Moreover, by providing not only global patient-level predictions but also task-specific model results that are related to radiological features, our pipeline aims to closely support the reading workflow of radiologists.
Soft Tissue Sarcoma Co-Segmentation in Combined MRI and PET/CT Data
Sep 24, 2020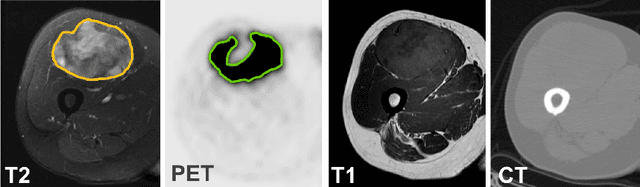
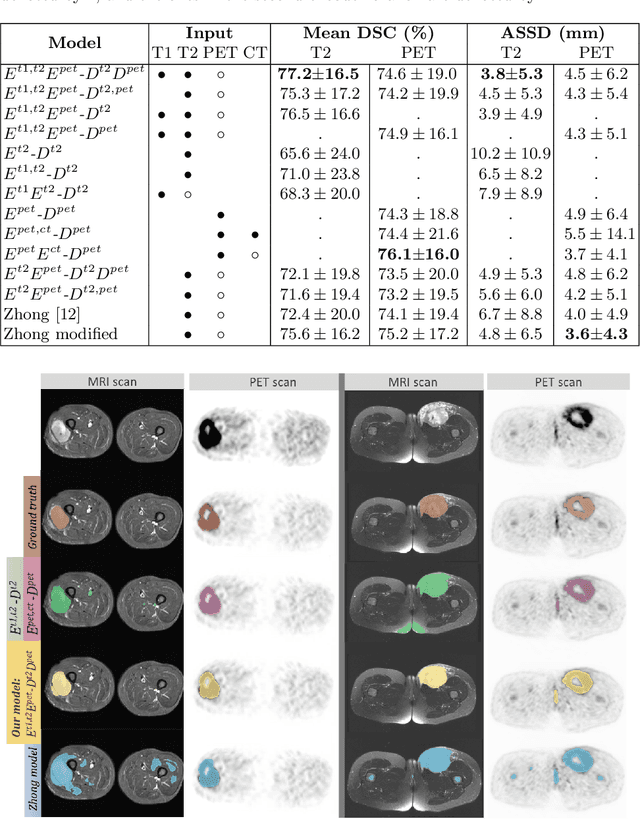
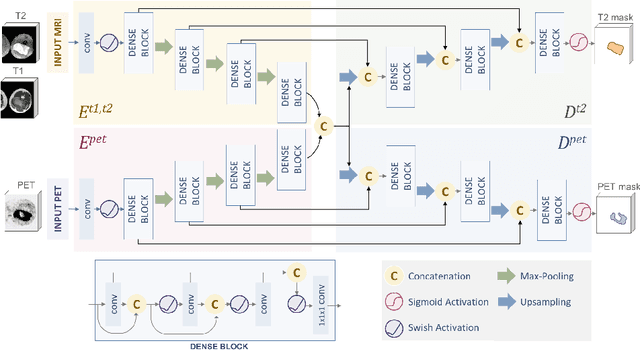
Abstract:Tumor segmentation in multimodal medical images has seen a growing trend towards deep learning based methods. Typically, studies dealing with this topic fuse multimodal image data to improve the tumor segmentation contour for a single imaging modality. However, they do not take into account that tumor characteristics are emphasized differently by each modality, which affects the tumor delineation. Thus, the tumor segmentation is modality- and task-dependent. This is especially the case for soft tissue sarcomas, where, due to necrotic tumor tissue, the segmentation differs vastly. Closing this gap, we develop a modalityspecific sarcoma segmentation model that utilizes multimodal image data to improve the tumor delineation on each individual modality. We propose a simultaneous co-segmentation method, which enables multimodal feature learning through modality-specific encoder and decoder branches, and the use of resource-effcient densely connected convolutional layers. We further conduct experiments to analyze how different input modalities and encoder-decoder fusion strategies affect the segmentation result. We demonstrate the effectiveness of our approach on public soft tissue sarcoma data, which comprises MRI (T1 and T2 sequence) and PET/CT scans. The results show that our multimodal co-segmentation model provides better modality-specific tumor segmentation than models using only the PET or MRI (T1 and T2) scan as input.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge