Annemiek Snoeckx
Employing similarity to highlight differences: On the impact of anatomical assumptions in chest X-ray registration methods
Jan 24, 2023



Abstract:To facilitate both the detection and the interpretation of findings in chest X-rays, comparison with a previous image of the same patient is very valuable to radiologists. Today, the most common approach for deep learning methods to automatically inspect chest X-rays disregards the patient history and classifies only single images as normal or abnormal. Nevertheless, several methods for assisting in the task of comparison through image registration have been proposed in the past. However, as we illustrate, they tend to miss specific types of pathological changes like cardiomegaly and effusion. Due to assumptions on fixed anatomical structures or their measurements of registration quality, they produce unnaturally deformed warp fields impacting visualization of differences between moving and fixed images. We aim to overcome these limitations, through a new paradigm based on individual rib pair segmentation for anatomy penalized registration. Our method proves to be a natural way to limit the folding percentage of the warp field to 1/6 of the state of the art while increasing the overlap of ribs by more than 25%, implying difference images showing pathological changes overlooked by other methods. We develop an anatomically penalized convolutional multi-stage solution on the National Institutes of Health (NIH) data set, starting from less than 25 fully and 50 partly labeled training images, employing sequential instance memory segmentation with hole dropout, weak labeling, coarse-to-fine refinement and Gaussian mixture model histogram matching. We statistically evaluate the benefits of our method and highlight the limits of currently used metrics for registration of chest X-rays.
Explainable-by-design Semi-Supervised Representation Learning for COVID-19 Diagnosis from CT Imaging
Dec 02, 2020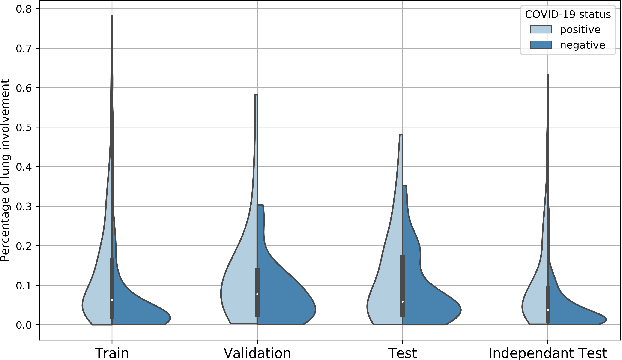
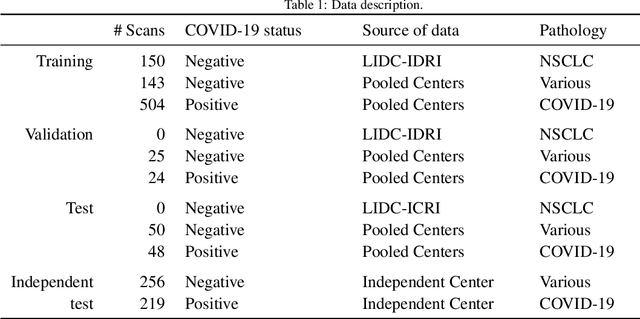
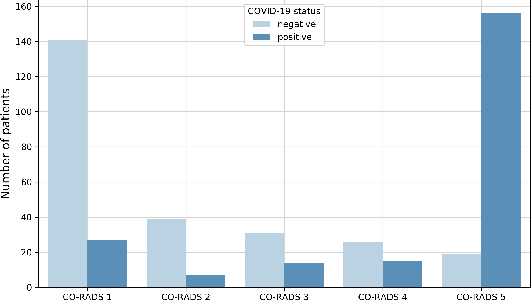
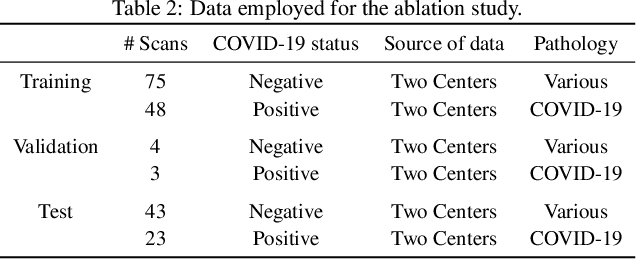
Abstract:Our motivating application is a real-world problem: COVID-19 classification from CT imaging, for which we present an explainable Deep Learning approach based on a semi-supervised classification pipeline that employs variational autoencoders to extract efficient feature embedding. We have optimized the architecture of two different networks for CT images: (i) a novel conditional variational autoencoder (CVAE) with a specific architecture that integrates the class labels inside the encoder layers and uses side information with shared attention layers for the encoder, which make the most of the contextual clues for representation learning, and (ii) a downstream convolutional neural network for supervised classification using the encoder structure of the CVAE. With the explainable classification results, the proposed diagnosis system is very effective for COVID-19 classification. Based on the promising results obtained qualitatively and quantitatively, we envisage a wide deployment of our developed technique in large-scale clinical studies.Code is available at https://git.etrovub.be/AVSP/ct-based-covid-19-diagnostic-tool.git.
Comparative study of deep learning methods for the automatic segmentation of lung, lesion and lesion type in CT scans of COVID-19 patients
Aug 21, 2020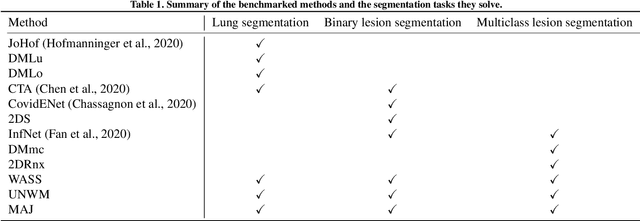
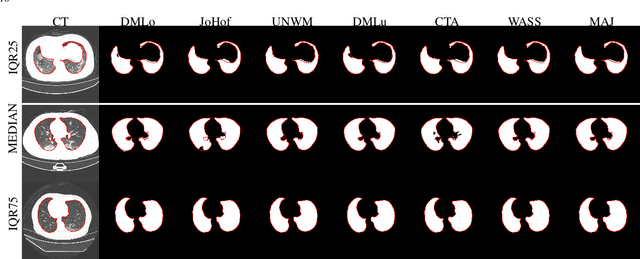
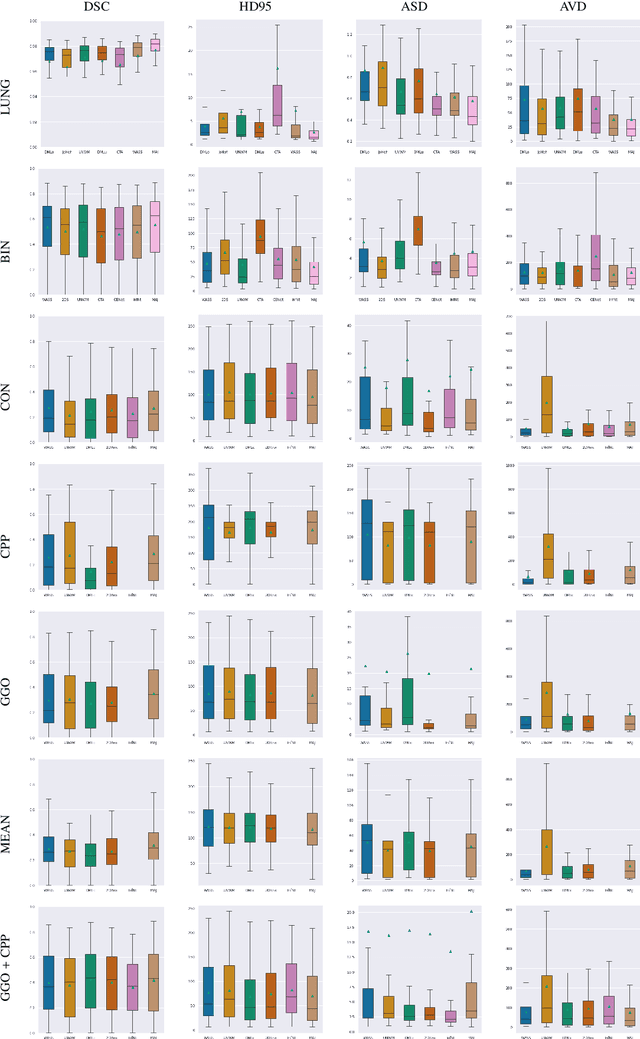
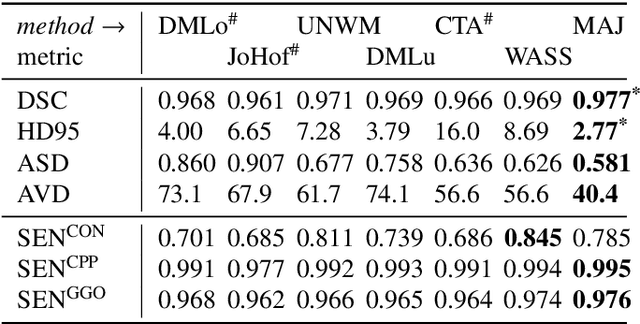
Abstract:Recent research on COVID-19 suggests that CT imaging provides useful information to assess disease progression and assist diagnosis, in addition to help understanding the disease. There is an increasing number of studies that propose to use deep learning to provide fast and accurate quantification of COVID-19 using chest CT scans. The main tasks of interest are the automatic segmentation of lung and lung lesions in chest CT scans of confirmed or suspected COVID-19 patients. In this study, we compare twelve deep learning algorithms using a multi-center dataset, including both open-source and in-house developed algorithms. Results show that ensembling different methods can boost the overall test set performance for lung segmentation, binary lesion segmentation and multiclass lesion segmentation, resulting in mean Dice scores of 0.982, 0.724 and 0.469, respectively. The resulting binary lesions were segmented with a mean absolute volume error of 91.3 ml. In general, the task of distinguishing different lesion types was more difficult, with a mean absolute volume difference of 152 ml and mean Dice scores of 0.369 and 0.523 for consolidation and ground glass opacity, respectively. All methods perform binary lesion segmentation with an average volume error that is better than visual assessment by human raters, suggesting these methods are mature enough for a large-scale evaluation for use in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge