Abel Díaz Berenguer
Representation Learning with Information Theory for COVID-19 Detection
Jul 04, 2022


Abstract:Successful data representation is a fundamental factor in machine learning based medical imaging analysis. Deep Learning (DL) has taken an essential role in robust representation learning. However, the inability of deep models to generalize to unseen data can quickly overfit intricate patterns. Thereby, we can conveniently implement strategies to aid deep models in discovering useful priors from data to learn their intrinsic properties. Our model, which we call a dual role network (DRN), uses a dependency maximization approach based on Least Squared Mutual Information (LSMI). The LSMI leverages dependency measures to ensure representation invariance and local smoothness. While prior works have used information theory measures like mutual information, known to be computationally expensive due to a density estimation step, our LSMI formulation alleviates the issues of intractable mutual information estimation and can be used to approximate it. Experiments on CT based COVID-19 Detection and COVID-19 Severity Detection benchmarks demonstrate the effectiveness of our method.
Explainable-by-design Semi-Supervised Representation Learning for COVID-19 Diagnosis from CT Imaging
Dec 02, 2020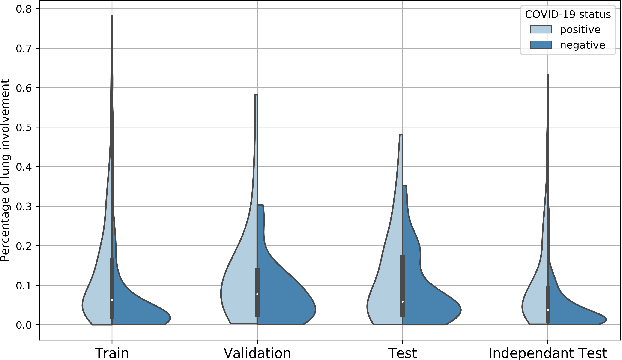
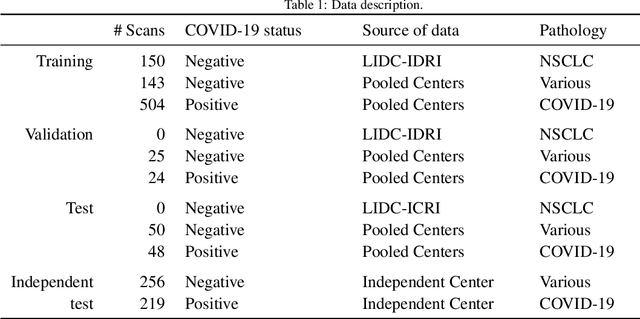
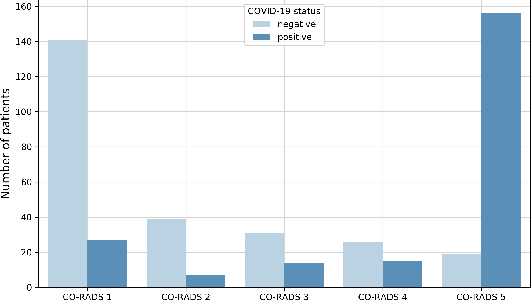
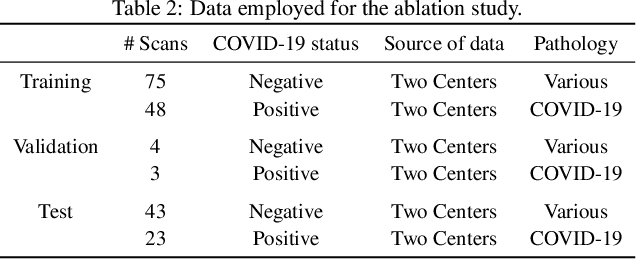
Abstract:Our motivating application is a real-world problem: COVID-19 classification from CT imaging, for which we present an explainable Deep Learning approach based on a semi-supervised classification pipeline that employs variational autoencoders to extract efficient feature embedding. We have optimized the architecture of two different networks for CT images: (i) a novel conditional variational autoencoder (CVAE) with a specific architecture that integrates the class labels inside the encoder layers and uses side information with shared attention layers for the encoder, which make the most of the contextual clues for representation learning, and (ii) a downstream convolutional neural network for supervised classification using the encoder structure of the CVAE. With the explainable classification results, the proposed diagnosis system is very effective for COVID-19 classification. Based on the promising results obtained qualitatively and quantitatively, we envisage a wide deployment of our developed technique in large-scale clinical studies.Code is available at https://git.etrovub.be/AVSP/ct-based-covid-19-diagnostic-tool.git.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge