Gert Sluiter
Multi-task fusion for improving mammography screening data classification
Dec 01, 2021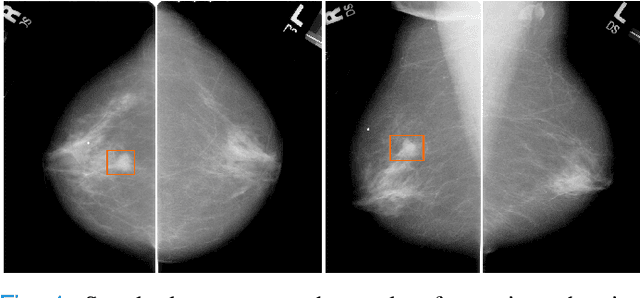
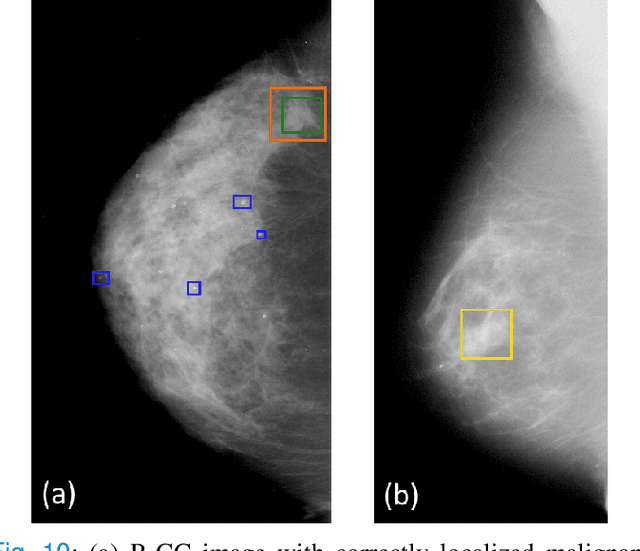
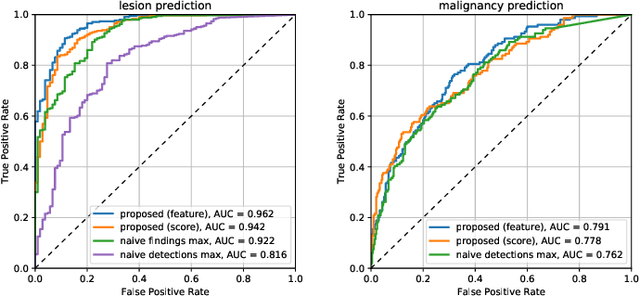
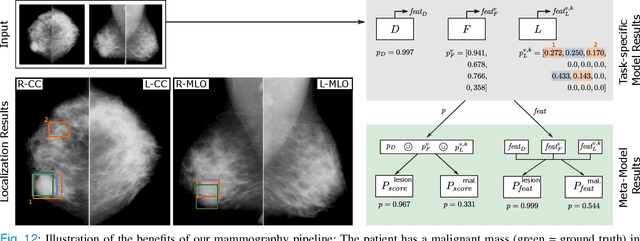
Abstract:Machine learning and deep learning methods have become essential for computer-assisted prediction in medicine, with a growing number of applications also in the field of mammography. Typically these algorithms are trained for a specific task, e.g., the classification of lesions or the prediction of a mammogram's pathology status. To obtain a comprehensive view of a patient, models which were all trained for the same task(s) are subsequently ensembled or combined. In this work, we propose a pipeline approach, where we first train a set of individual, task-specific models and subsequently investigate the fusion thereof, which is in contrast to the standard model ensembling strategy. We fuse model predictions and high-level features from deep learning models with hybrid patient models to build stronger predictors on patient level. To this end, we propose a multi-branch deep learning model which efficiently fuses features across different tasks and mammograms to obtain a comprehensive patient-level prediction. We train and evaluate our full pipeline on public mammography data, i.e., DDSM and its curated version CBIS-DDSM, and report an AUC score of 0.962 for predicting the presence of any lesion and 0.791 for predicting the presence of malignant lesions on patient level. Overall, our fusion approaches improve AUC scores significantly by up to 0.04 compared to standard model ensembling. Moreover, by providing not only global patient-level predictions but also task-specific model results that are related to radiological features, our pipeline aims to closely support the reading workflow of radiologists.
Domain aware medical image classifier interpretation by counterfactual impact analysis
Jul 13, 2020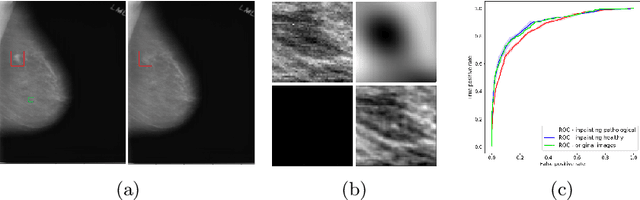
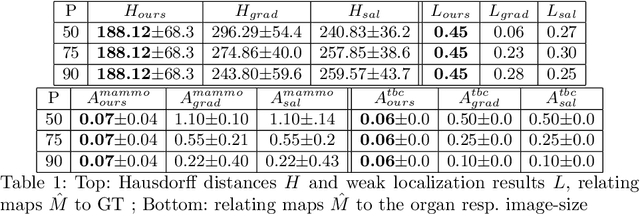
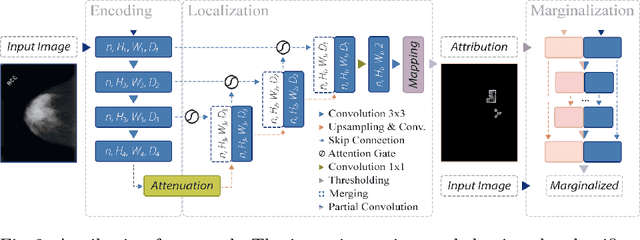
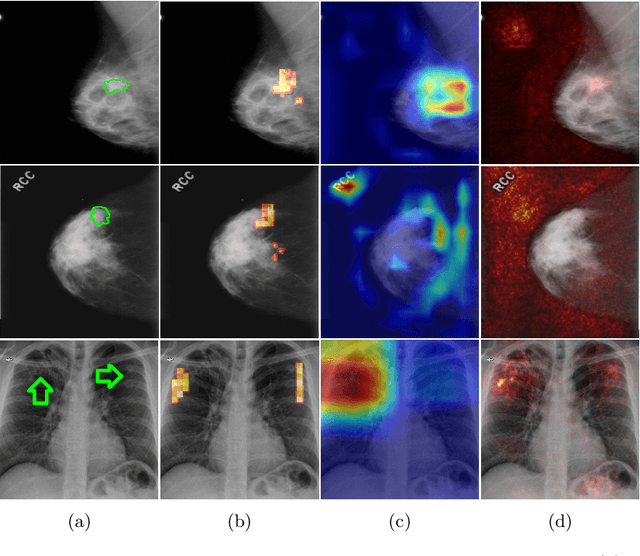
Abstract:The success of machine learning methods for computer vision tasks has driven a surge in computer assisted prediction for medicine and biology. Based on a data-driven relationship between input image and pathological classification, these predictors deliver unprecedented accuracy. Yet, the numerous approaches trying to explain the causality of this learned relationship have fallen short: time constraints, coarse, diffuse and at times misleading results, caused by the employment of heuristic techniques like Gaussian noise and blurring, have hindered their clinical adoption. In this work, we discuss and overcome these obstacles by introducing a neural-network based attribution method, applicable to any trained predictor. Our solution identifies salient regions of an input image in a single forward-pass by measuring the effect of local image-perturbations on a predictor's score. We replace heuristic techniques with a strong neighborhood conditioned inpainting approach, avoiding anatomically implausible, hence adversarial artifacts. We evaluate on public mammography data and compare against existing state-of-the-art methods. Furthermore, we exemplify the approach's generalizability by demonstrating results on chest X-rays. Our solution shows, both quantitatively and qualitatively, a significant reduction of localization ambiguity and clearer conveying results, without sacrificing time efficiency.
Interpreting Medical Image Classifiers by Optimization Based Counterfactual Impact Analysis
Apr 03, 2020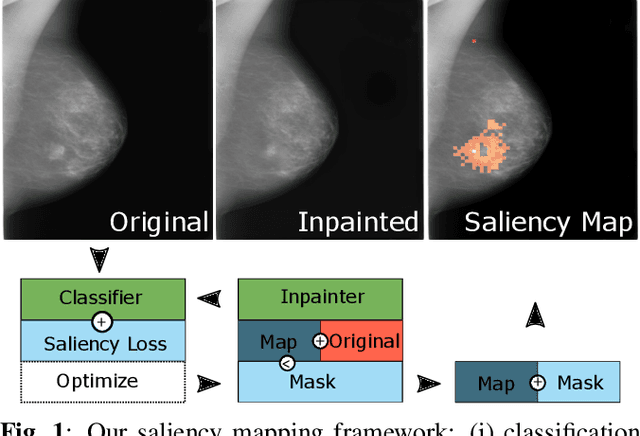
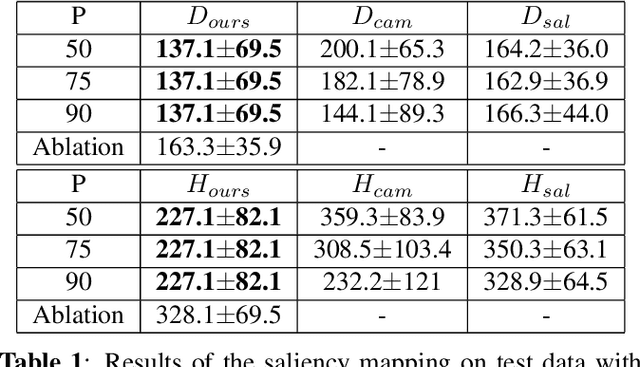
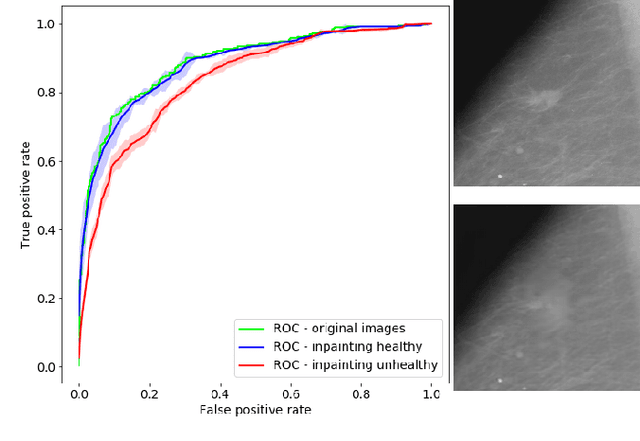
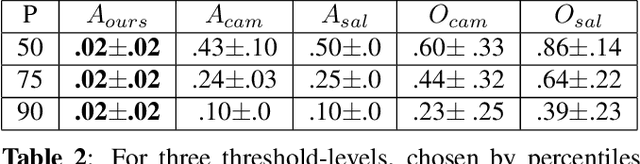
Abstract:Clinical applicability of automated decision support systems depends on a robust, well-understood classification interpretation. Artificial neural networks while achieving class-leading scores fall short in this regard. Therefore, numerous approaches have been proposed that map a salient region of an image to a diagnostic classification. Utilizing heuristic methodology, like blurring and noise, they tend to produce diffuse, sometimes misleading results, hindering their general adoption. In this work we overcome these issues by presenting a model agnostic saliency mapping framework tailored to medical imaging. We replace heuristic techniques with a strong neighborhood conditioned inpainting approach, which avoids anatomically implausible artefacts. We formulate saliency attribution as a map-quality optimization task, enforcing constrained and focused attributions. Experiments on public mammography data show quantitatively and qualitatively more precise localization and clearer conveying results than existing state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge