Robert Toth
Toth Technology LLC, Dover, NJ, USA
PatchSorter: A High Throughput Deep Learning Digital Pathology Tool for Object Labeling
Jul 13, 2023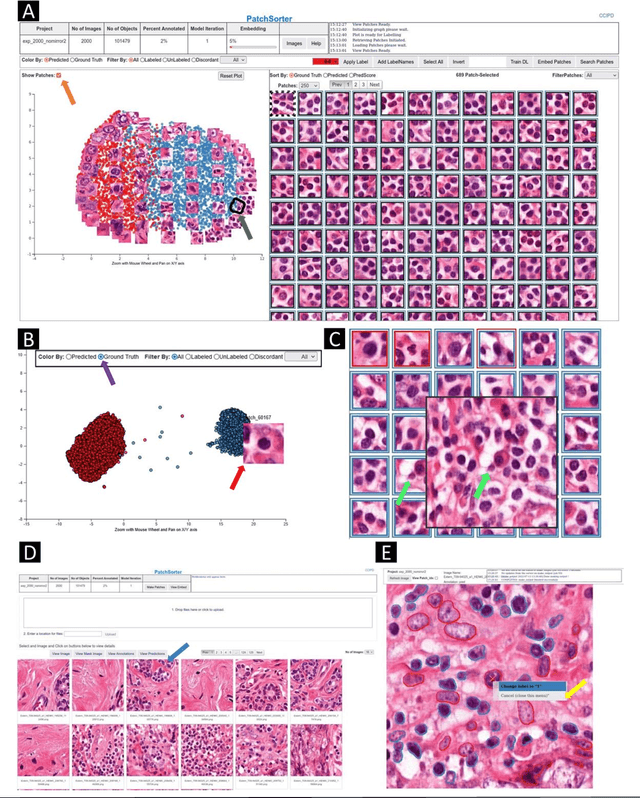
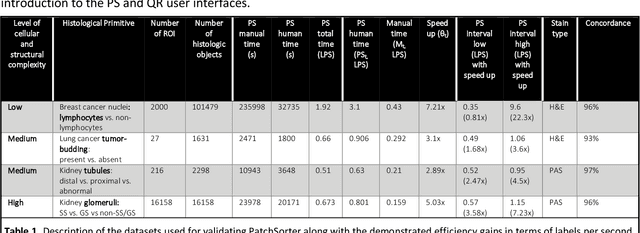
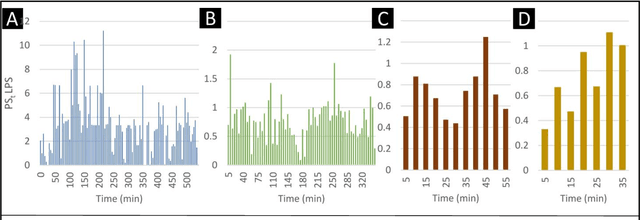
Abstract:The discovery of patterns associated with diagnosis, prognosis, and therapy response in digital pathology images often requires intractable labeling of large quantities of histological objects. Here we release an open-source labeling tool, PatchSorter, which integrates deep learning with an intuitive web interface. Using >100,000 objects, we demonstrate a >7x improvement in labels per second over unaided labeling, with minimal impact on labeling accuracy, thus enabling high-throughput labeling of large datasets.
Computationally efficient neural network classifiers for next generation closed loop neuromodulation therapy, a case study in epilepsy
Apr 27, 2022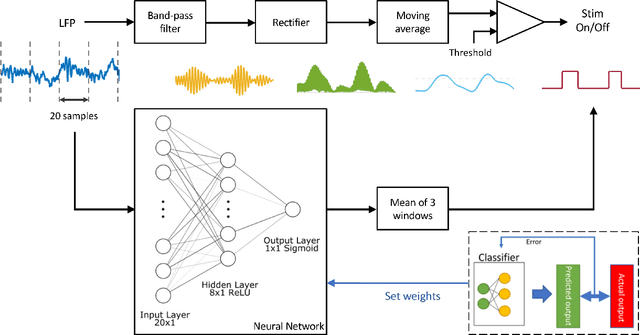
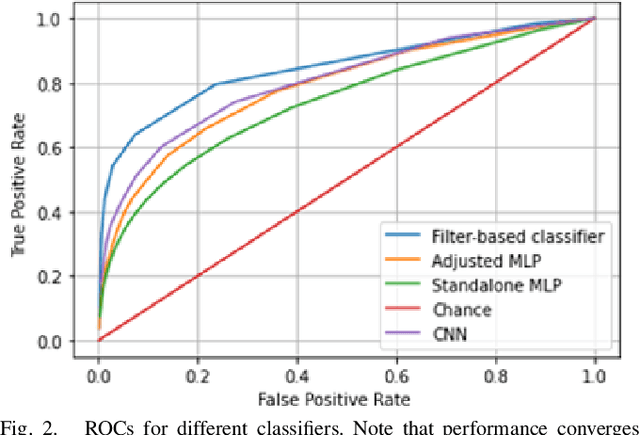
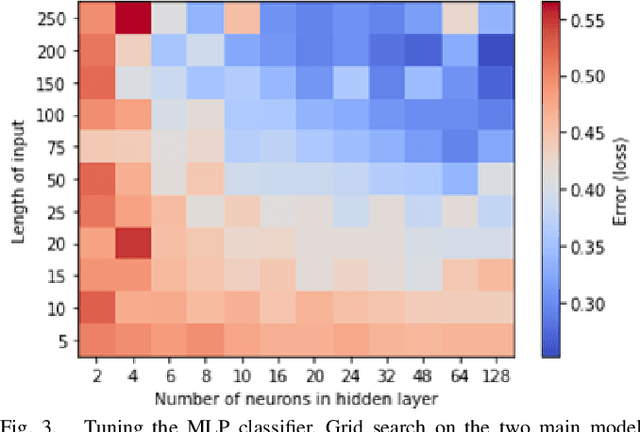
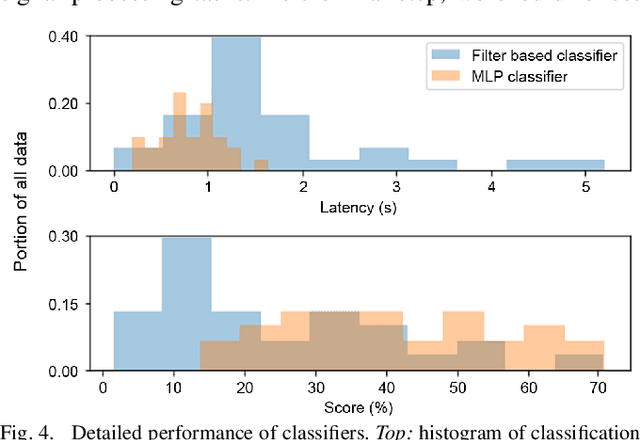
Abstract:This work explores the potential utility of neural network classifiers for real-time classification of field-potential based biomarkers in next-generation responsive neuromodulation systems. Compared to classical filter-based classifiers, neural networks offer an ease of patient-specific parameter tuning, promising to reduce the burden of programming on clinicians. The paper explores a compact, feed-forward neural network architecture of only dozens of units for seizure-state classification in refractory epilepsy. The proposed classifier offers comparable accuracy to filter classifiers on clinician-labelled data, while reducing detection latency. As a trade-off to classical methods, the paper focuses on keeping the complexity of the architecture minimal, to accommodate the on-board computational constraints of implantable pulse generator systems.
Embedding digital chronotherapy into medical devices -- A canine case study in controlling status epilepticus through multi-scale rhythmic brain stimulation
Jul 07, 2021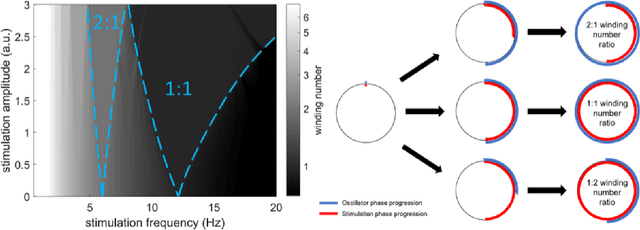
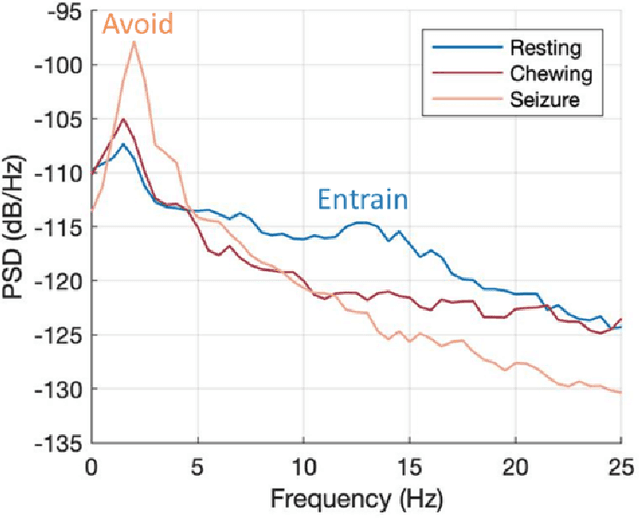
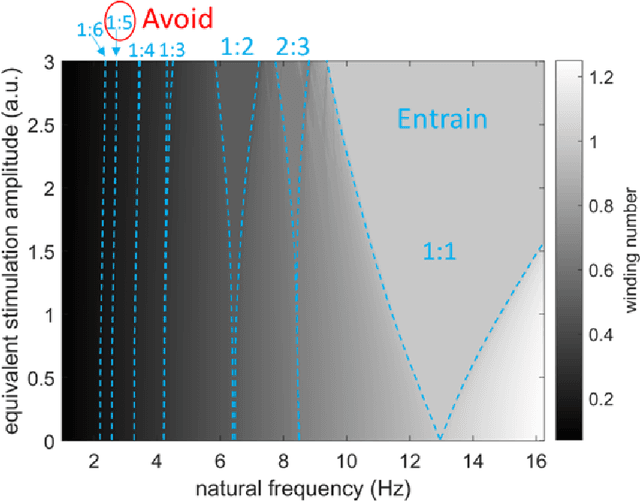

Abstract:Circadian and other physiological rhythms play a key role in both normal homeostasis and disease processes. Such is the case of circadian and infradian seizure patterns observed in epilepsy. However, these rhythms are not fully exploited in the design of active implantable medical devices. In this paper we explore a new implantable stimulator that implements chronotherapy as a feedforward input to supplement both open-loop and closed-loop methods. This integrated algorithm allows for stimulation to be adjusted to the ultradian, circadian, and infradian patterns observed in patients through slowly-varying temporal adjustments of stimulation and algorithm sub-components, while also enabling adaption of stimulation based on immediate physiological needs such as a breakthrough seizure or change of posture. Embedded physiological sensors in the stimulator can be used to refine the baseline stimulation circadian pattern as a "digital zeitgeber". This algorithmic approach is tested on a canine with severe drug-resistant idiopathic generalized epilepsy exhibiting a characteristic diurnal pattern correlated with sleep-wake cycles. Prior to implantation, the canine's cluster seizures evolved to status epilepticus (SE) and required emergency pharmacological intervention. The cranially-mounted system was fully-implanted bilaterally into the centromedian nucleus of the thalamus. Using combinations of time-based modulation, thalamocortical rhythm-specific tuning of frequency parameters, and fast-adaptive modes based on activity, the canine has experienced no further SE events post-implant at the time of writing (7 months), and no significant clusters are observed any longer. The use of digitally-enabled chronotherapy as a feedforward signal to augment adaptive neurostimulators could prove a useful algorithmic method where sensitivity to temporal patterns are characteristics of the disease state.
Quick Annotator: an open-source digital pathology based rapid image annotation tool
Jan 06, 2021



Abstract:Image based biomarker discovery typically requires an accurate segmentation of histologic structures (e.g., cell nuclei, tubules, epithelial regions) in digital pathology Whole Slide Images (WSI). Unfortunately, annotating each structure of interest is laborious and often intractable even in moderately sized cohorts. Here, we present an open-source tool, Quick Annotator (QA), designed to improve annotation efficiency of histologic structures by orders of magnitude. While the user annotates regions of interest (ROI) via an intuitive web interface, a deep learning (DL) model is concurrently optimized using these annotations and applied to the ROI. The user iteratively reviews DL results to either (a) accept accurately annotated regions, or (b) correct erroneously segmented structures to improve subsequent model suggestions, before transitioning to other ROIs. We demonstrate the effectiveness of QA over comparable manual efforts via three use cases. These include annotating (a) 337,386 nuclei in 5 pancreatic WSIs, (b) 5,692 tubules in 10 colorectal WSIs, and (c) 14,187 regions of epithelium in 10 breast WSIs. Efficiency gains in terms of annotations per second of 102x, 9x, and 39x were respectively witnessed while retaining f-scores >.95, suggesting QA may be a valuable tool for efficiently fully annotating WSIs employed in downstream biomarker studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge