Timothy Denison
BioX-Bridge: Model Bridging for Unsupervised Cross-Modal Knowledge Transfer across Biosignals
Oct 02, 2025Abstract:Biosignals offer valuable insights into the physiological states of the human body. Although biosignal modalities differ in functionality, signal fidelity, sensor comfort, and cost, they are often intercorrelated, reflecting the holistic and interconnected nature of human physiology. This opens up the possibility of performing the same tasks using alternative biosignal modalities, thereby improving the accessibility, usability, and adaptability of health monitoring systems. However, the limited availability of large labeled datasets presents challenges for training models tailored to specific tasks and modalities of interest. Unsupervised cross-modal knowledge transfer offers a promising solution by leveraging knowledge from an existing modality to support model training for a new modality. Existing methods are typically based on knowledge distillation, which requires running a teacher model alongside student model training, resulting in high computational and memory overhead. This challenge is further exacerbated by the recent development of foundation models that demonstrate superior performance and generalization across tasks at the cost of large model sizes. To this end, we explore a new framework for unsupervised cross-modal knowledge transfer of biosignals by training a lightweight bridge network to align the intermediate representations and enable information flow between foundation models and across modalities. Specifically, we introduce an efficient strategy for selecting alignment positions where the bridge should be constructed, along with a flexible prototype network as the bridge architecture. Extensive experiments across multiple biosignal modalities, tasks, and datasets show that BioX-Bridge reduces the number of trainable parameters by 88--99\% while maintaining or even improving transfer performance compared to state-of-the-art methods.
AnchorInv: Few-Shot Class-Incremental Learning of Physiological Signals via Representation Space Guided Inversion
Dec 18, 2024



Abstract:Deep learning models have demonstrated exceptional performance in a variety of real-world applications. These successes are often attributed to strong base models that can generalize to novel tasks with limited supporting data while keeping prior knowledge intact. However, these impressive results are based on the availability of a large amount of high-quality data, which is often lacking in specialized biomedical applications. In such fields, models are usually developed with limited data that arrive incrementally with novel categories. This requires the model to adapt to new information while preserving existing knowledge. Few-Shot Class-Incremental Learning (FSCIL) methods offer a promising approach to addressing these challenges, but they also depend on strong base models that face the same aforementioned limitations. To overcome these constraints, we propose AnchorInv following the straightforward and efficient buffer-replay strategy. Instead of selecting and storing raw data, AnchorInv generates synthetic samples guided by anchor points in the feature space. This approach protects privacy and regularizes the model for adaptation. When evaluated on three public physiological time series datasets, AnchorInv exhibits efficient knowledge forgetting prevention and improved adaptation to novel classes, surpassing state-of-the-art baselines.
A Survey of Few-Shot Learning for Biomedical Time Series
May 03, 2024



Abstract:Advancements in wearable sensor technologies and the digitization of medical records have contributed to the unprecedented ubiquity of biomedical time series data. Data-driven models have tremendous potential to assist clinical diagnosis and improve patient care by improving long-term monitoring capabilities, facilitating early disease detection and intervention, as well as promoting personalized healthcare delivery. However, accessing extensively labeled datasets to train data-hungry deep learning models encounters many barriers, such as long-tail distribution of rare diseases, cost of annotation, privacy and security concerns, data-sharing regulations, and ethical considerations. An emerging approach to overcome the scarcity of labeled data is to augment AI methods with human-like capabilities to leverage past experiences to learn new tasks with limited examples, called few-shot learning. This survey provides a comprehensive review and comparison of few-shot learning methods for biomedical time series applications. The clinical benefits and limitations of such methods are discussed in relation to traditional data-driven approaches. This paper aims to provide insights into the current landscape of few-shot learning for biomedical time series and its implications for future research and applications.
MorpheusNet: Resource efficient sleep stage classifier for embedded on-line systems
Jan 14, 2024
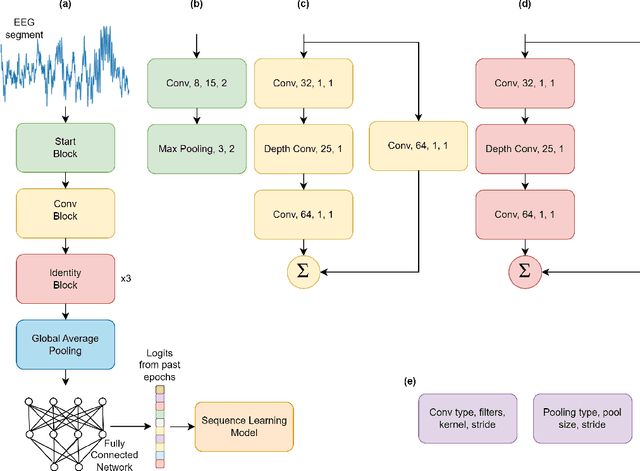


Abstract:Sleep Stage Classification (SSC) is a labor-intensive task, requiring experts to examine hours of electrophysiological recordings for manual classification. This is a limiting factor when it comes to leveraging sleep stages for therapeutic purposes. With increasing affordability and expansion of wearable devices, automating SSC may enable deployment of sleep-based therapies at scale. Deep Learning has gained increasing attention as a potential method to automate this process. Previous research has shown accuracy comparable to manual expert scores. However, previous approaches require sizable amount of memory and computational resources. This constrains the ability to classify in real time and deploy models on the edge. To address this gap, we aim to provide a model capable of predicting sleep stages in real-time, without requiring access to external computational sources (e.g., mobile phone, cloud). The algorithm is power efficient to enable use on embedded battery powered systems. Our compact sleep stage classifier can be deployed on most off-the-shelf microcontrollers (MCU) with constrained hardware settings. This is due to the memory footprint of our approach requiring significantly fewer operations. The model was tested on three publicly available data bases and achieved performance comparable to the state of the art, whilst reducing model complexity by orders of magnitude (up to 280 times smaller compared to state of the art). We further optimized the model with quantization of parameters to 8 bits with only an average drop of 0.95% in accuracy. When implemented in firmware, the quantized model achieves a latency of 1.6 seconds on an Arm CortexM4 processor, allowing its use for on-line SSC-based therapies.
Computationally efficient neural network classifiers for next generation closed loop neuromodulation therapy, a case study in epilepsy
Apr 27, 2022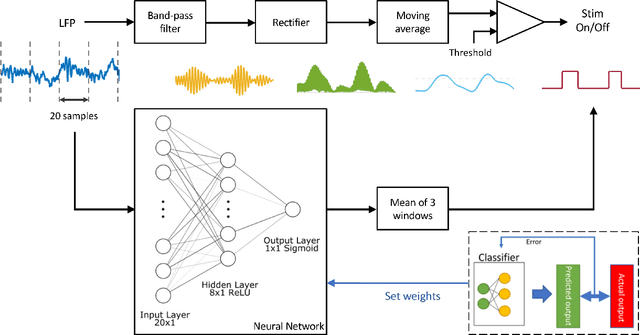
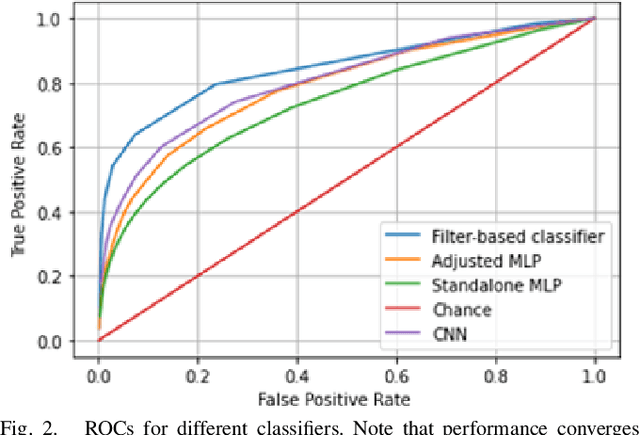
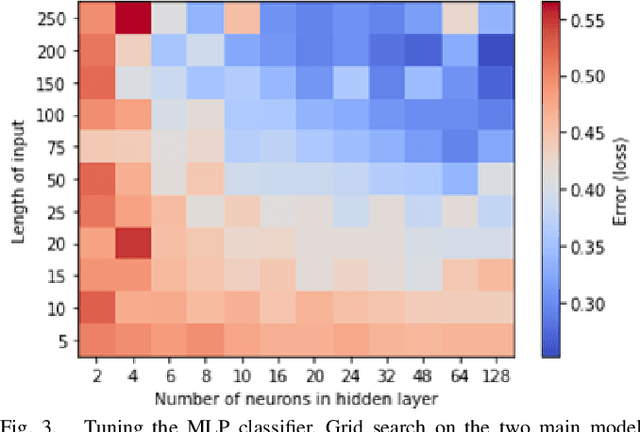
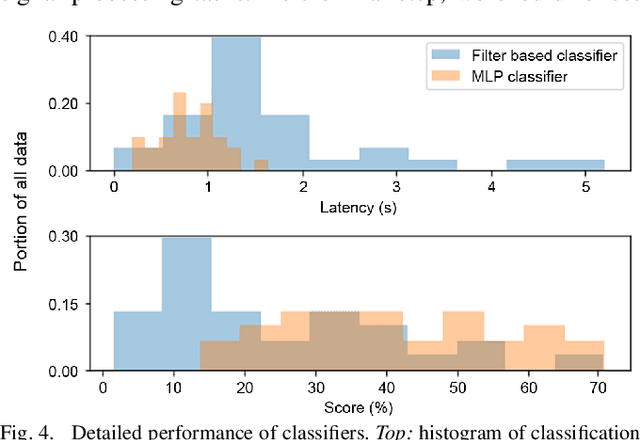
Abstract:This work explores the potential utility of neural network classifiers for real-time classification of field-potential based biomarkers in next-generation responsive neuromodulation systems. Compared to classical filter-based classifiers, neural networks offer an ease of patient-specific parameter tuning, promising to reduce the burden of programming on clinicians. The paper explores a compact, feed-forward neural network architecture of only dozens of units for seizure-state classification in refractory epilepsy. The proposed classifier offers comparable accuracy to filter classifiers on clinician-labelled data, while reducing detection latency. As a trade-off to classical methods, the paper focuses on keeping the complexity of the architecture minimal, to accommodate the on-board computational constraints of implantable pulse generator systems.
Embedding digital chronotherapy into medical devices -- A canine case study in controlling status epilepticus through multi-scale rhythmic brain stimulation
Jul 07, 2021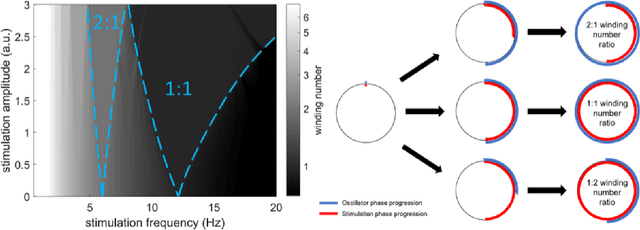
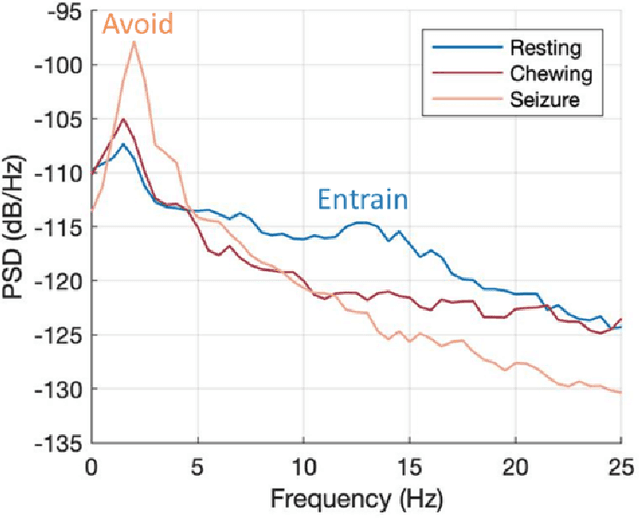
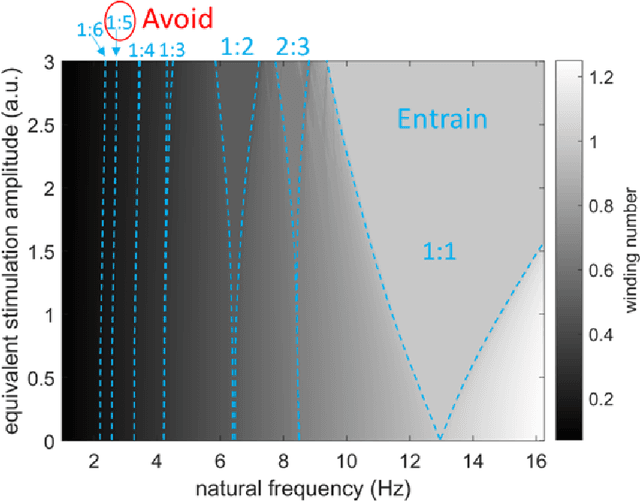

Abstract:Circadian and other physiological rhythms play a key role in both normal homeostasis and disease processes. Such is the case of circadian and infradian seizure patterns observed in epilepsy. However, these rhythms are not fully exploited in the design of active implantable medical devices. In this paper we explore a new implantable stimulator that implements chronotherapy as a feedforward input to supplement both open-loop and closed-loop methods. This integrated algorithm allows for stimulation to be adjusted to the ultradian, circadian, and infradian patterns observed in patients through slowly-varying temporal adjustments of stimulation and algorithm sub-components, while also enabling adaption of stimulation based on immediate physiological needs such as a breakthrough seizure or change of posture. Embedded physiological sensors in the stimulator can be used to refine the baseline stimulation circadian pattern as a "digital zeitgeber". This algorithmic approach is tested on a canine with severe drug-resistant idiopathic generalized epilepsy exhibiting a characteristic diurnal pattern correlated with sleep-wake cycles. Prior to implantation, the canine's cluster seizures evolved to status epilepticus (SE) and required emergency pharmacological intervention. The cranially-mounted system was fully-implanted bilaterally into the centromedian nucleus of the thalamus. Using combinations of time-based modulation, thalamocortical rhythm-specific tuning of frequency parameters, and fast-adaptive modes based on activity, the canine has experienced no further SE events post-implant at the time of writing (7 months), and no significant clusters are observed any longer. The use of digitally-enabled chronotherapy as a feedforward signal to augment adaptive neurostimulators could prove a useful algorithmic method where sensitivity to temporal patterns are characteristics of the disease state.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge