Evan Mason
Can tumor location on pre-treatment MRI predict likelihood of pseudo-progression versus tumor recurrence in Glioblastoma? A feasibility study
Jun 16, 2020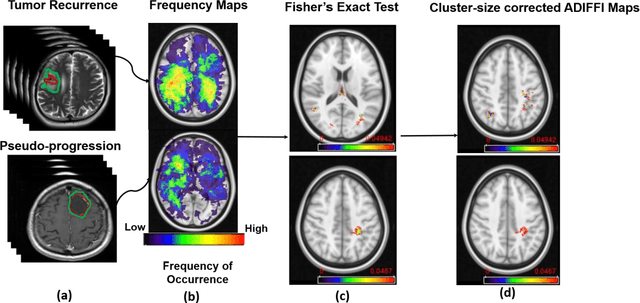

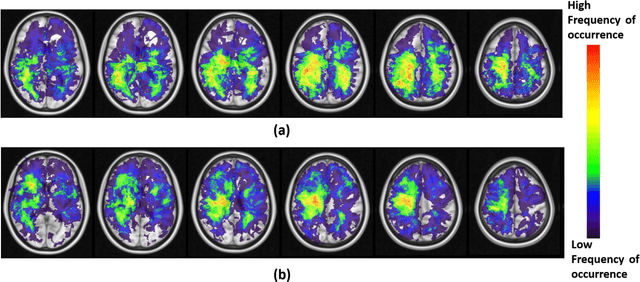
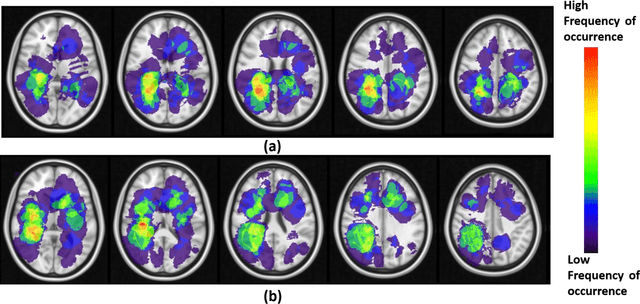
Abstract:A significant challenge in Glioblastoma (GBM) management is identifying pseudo-progression (PsP), a benign radiation-induced effect, from tumor recurrence, on routine imaging following conventional treatment. Previous studies have linked tumor lobar presence and laterality to GBM outcomes, suggesting that disease etiology and progression in GBM may be impacted by tumor location. Hence, in this feasibility study, we seek to investigate the following question: Can tumor location on treatment-na\"ive MRI provide early cues regarding likelihood of a patient developing pseudo-progression versus tumor recurrence? In this study, 74 pre-treatment Glioblastoma MRI scans with PsP (33) and tumor recurrence (41) were analyzed. First, enhancing lesion on Gd-T1w MRI and peri-lesional hyperintensities on T2w/FLAIR were segmented by experts and then registered to a brain atlas. Using patients from the two phenotypes, we construct two atlases by quantifying frequency of occurrence of enhancing lesion and peri-lesion hyperintensities, by averaging voxel intensities across the population. Analysis of differential involvement was then performed to compute voxel-wise significant differences (p-value<0.05) across the atlases. Statistically significant clusters were finally mapped to a structural atlas to provide anatomic localization of their location. Our results demonstrate that patients with tumor recurrence showed prominence of their initial tumor in the parietal lobe, while patients with PsP showed a multi-focal distribution of the initial tumor in the frontal and temporal lobes, insula, and putamen. These preliminary results suggest that lateralization of pre-treatment lesions towards certain anatomical areas of the brain may allow to provide early cues regarding assessing likelihood of occurrence of pseudo-progression from tumor recurrence on MRI scans.
EddyNet: A Deep Neural Network For Pixel-Wise Classification of Oceanic Eddies
Nov 10, 2017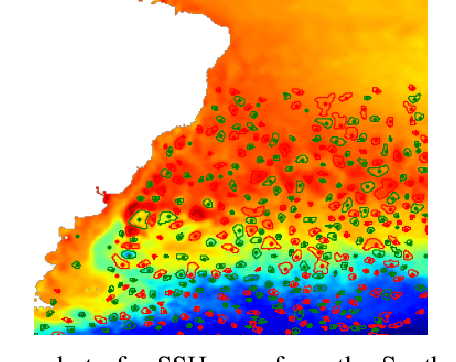
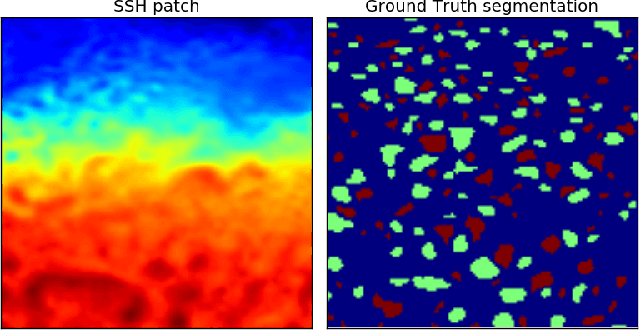
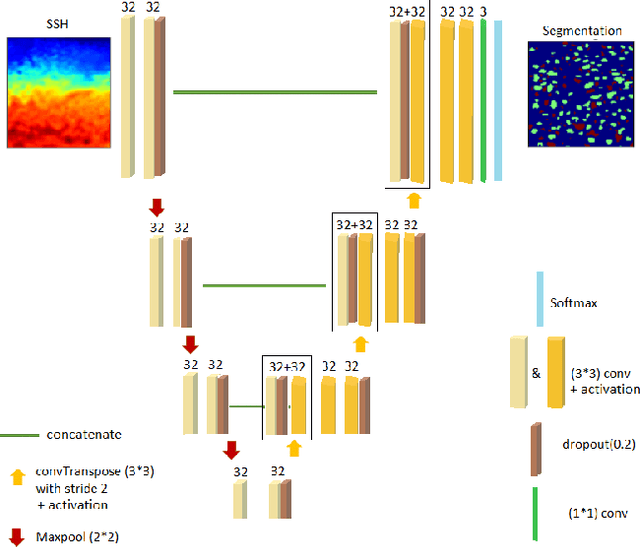
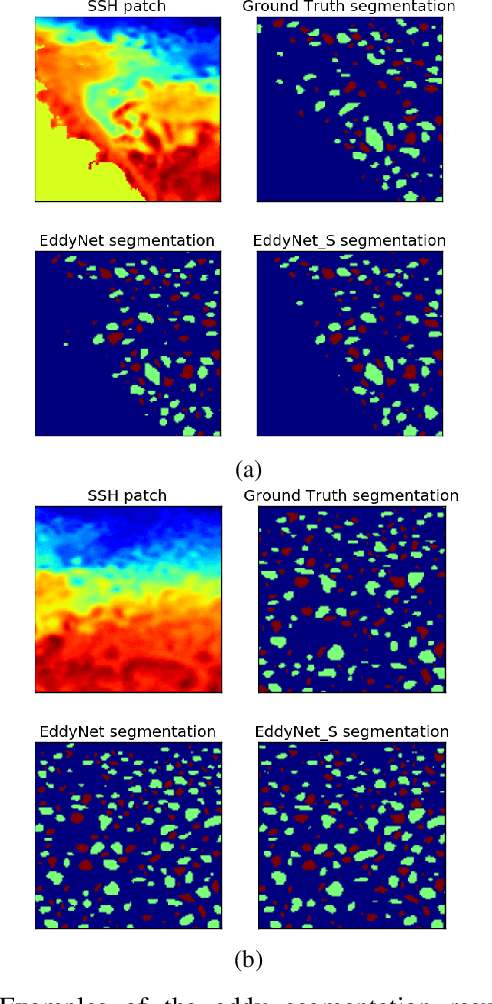
Abstract:This work presents EddyNet, a deep learning based architecture for automated eddy detection and classification from Sea Surface Height (SSH) maps provided by the Copernicus Marine and Environment Monitoring Service (CMEMS). EddyNet is a U-Net like network that consists of a convolutional encoder-decoder followed by a pixel-wise classification layer. The output is a map with the same size of the input where pixels have the following labels \{'0': Non eddy, '1': anticyclonic eddy, '2': cyclonic eddy\}. We investigate the use of SELU activation function instead of the classical ReLU+BN and we use an overlap based loss function instead of the cross entropy loss. Keras Python code, the training datasets and EddyNet weights files are open-source and freely available on https://github.com/redouanelg/EddyNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge