Monica Munnangi
Assessing the Limitations of Large Language Models in Clinical Fact Decomposition
Dec 17, 2024Abstract:Verifying factual claims is critical for using large language models (LLMs) in healthcare. Recent work has proposed fact decomposition, which uses LLMs to rewrite source text into concise sentences conveying a single piece of information, as an approach for fine-grained fact verification. Clinical documentation poses unique challenges for fact decomposition due to dense terminology and diverse note types. To explore these challenges, we present FactEHR, a dataset consisting of full document fact decompositions for 2,168 clinical notes spanning four types from three hospital systems. Our evaluation, including review by clinicians, highlights significant variability in the quality of fact decomposition for four commonly used LLMs, with some LLMs generating 2.6x more facts per sentence than others. The results underscore the need for better LLM capabilities to support factual verification in clinical text. To facilitate future research in this direction, we plan to release our code at \url{https://github.com/som-shahlab/factehr}.
A Brief History of Named Entity Recognition
Nov 07, 2024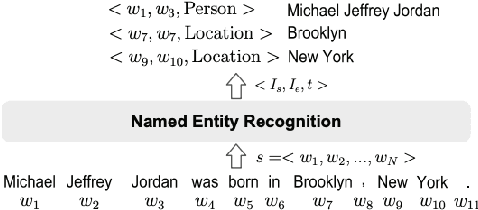

Abstract:A large amount of information in today's world is now stored in knowledge bases. Named Entity Recognition (NER) is a process of extracting, disambiguation, and linking an entity from raw text to insightful and structured knowledge bases. More concretely, it is identifying and classifying entities in the text that are crucial for Information Extraction, Semantic Annotation, Question Answering, Ontology Population, and so on. The process of NER has evolved in the last three decades since it first appeared in 1996. In this survey, we study the evolution of techniques employed for NER and compare the results, starting from supervised to the developing unsupervised learning methods.
Open (Clinical) LLMs are Sensitive to Instruction Phrasings
Jul 12, 2024



Abstract:Instruction-tuned Large Language Models (LLMs) can perform a wide range of tasks given natural language instructions to do so, but they are sensitive to how such instructions are phrased. This issue is especially concerning in healthcare, as clinicians are unlikely to be experienced prompt engineers and the potential consequences of inaccurate outputs are heightened in this domain. This raises a practical question: How robust are instruction-tuned LLMs to natural variations in the instructions provided for clinical NLP tasks? We collect prompts from medical doctors across a range of tasks and quantify the sensitivity of seven LLMs -- some general, others specialized -- to natural (i.e., non-adversarial) instruction phrasings. We find that performance varies substantially across all models, and that -- perhaps surprisingly -- domain-specific models explicitly trained on clinical data are especially brittle, compared to their general domain counterparts. Further, arbitrary phrasing differences can affect fairness, e.g., valid but distinct instructions for mortality prediction yield a range both in overall performance, and in terms of differences between demographic groups.
On-the-fly Definition Augmentation of LLMs for Biomedical NER
Mar 29, 2024Abstract:Despite their general capabilities, LLMs still struggle on biomedical NER tasks, which are difficult due to the presence of specialized terminology and lack of training data. In this work we set out to improve LLM performance on biomedical NER in limited data settings via a new knowledge augmentation approach which incorporates definitions of relevant concepts on-the-fly. During this process, to provide a test bed for knowledge augmentation, we perform a comprehensive exploration of prompting strategies. Our experiments show that definition augmentation is useful for both open source and closed LLMs. For example, it leads to a relative improvement of 15\% (on average) in GPT-4 performance (F1) across all (six) of our test datasets. We conduct extensive ablations and analyses to demonstrate that our performance improvements stem from adding relevant definitional knowledge. We find that careful prompting strategies also improve LLM performance, allowing them to outperform fine-tuned language models in few-shot settings. To facilitate future research in this direction, we release our code at https://github.com/allenai/beacon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge