Silvio Amir
Elucidating Mechanisms of Demographic Bias in LLMs for Healthcare
Feb 18, 2025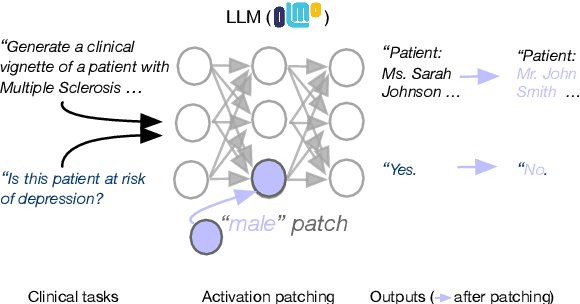

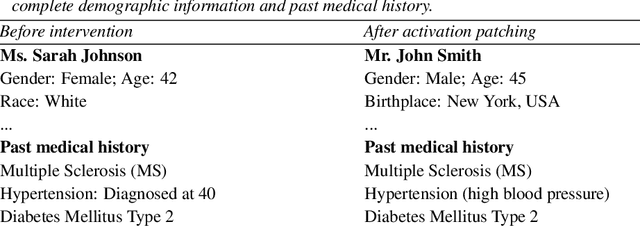
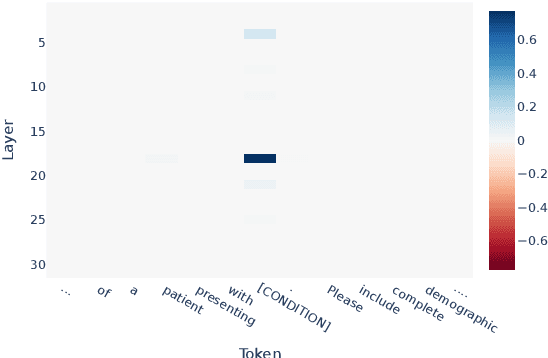
Abstract:We know from prior work that LLMs encode social biases, and that this manifests in clinical tasks. In this work we adopt tools from mechanistic interpretability to unveil sociodemographic representations and biases within LLMs in the context of healthcare. Specifically, we ask: Can we identify activations within LLMs that encode sociodemographic information (e.g., gender, race)? We find that gender information is highly localized in middle MLP layers and can be reliably manipulated at inference time via patching. Such interventions can surgically alter generated clinical vignettes for specific conditions, and also influence downstream clinical predictions which correlate with gender, e.g., patient risk of depression. We find that representation of patient race is somewhat more distributed, but can also be intervened upon, to a degree. To our knowledge, this is the first application of mechanistic interpretability methods to LLMs for healthcare.
Who Taught You That? Tracing Teachers in Model Distillation
Feb 10, 2025Abstract:Model distillation -- using outputs from a large teacher model to teach a small student model -- is a practical means of creating efficient models for a particular task. We ask: Can we identify a students' teacher based on its outputs? Such "footprints" left by teacher LLMs would be interesting artifacts. Beyond this, reliable teacher inference may have practical implications as actors seek to distill specific capabilities of massive proprietary LLMs into deployed smaller LMs, potentially violating terms of service. We consider practical task distillation targets including summarization, question answering, and instruction-following. We assume a finite set of candidate teacher models, which we treat as blackboxes. We design discriminative models that operate over lexical features. We find that $n$-gram similarity alone is unreliable for identifying teachers, but part-of-speech (PoS) templates preferred by student models mimic those of their teachers.
Open (Clinical) LLMs are Sensitive to Instruction Phrasings
Jul 12, 2024



Abstract:Instruction-tuned Large Language Models (LLMs) can perform a wide range of tasks given natural language instructions to do so, but they are sensitive to how such instructions are phrased. This issue is especially concerning in healthcare, as clinicians are unlikely to be experienced prompt engineers and the potential consequences of inaccurate outputs are heightened in this domain. This raises a practical question: How robust are instruction-tuned LLMs to natural variations in the instructions provided for clinical NLP tasks? We collect prompts from medical doctors across a range of tasks and quantify the sensitivity of seven LLMs -- some general, others specialized -- to natural (i.e., non-adversarial) instruction phrasings. We find that performance varies substantially across all models, and that -- perhaps surprisingly -- domain-specific models explicitly trained on clinical data are especially brittle, compared to their general domain counterparts. Further, arbitrary phrasing differences can affect fairness, e.g., valid but distinct instructions for mortality prediction yield a range both in overall performance, and in terms of differences between demographic groups.
Investigating Mysteries of CoT-Augmented Distillation
Jun 20, 2024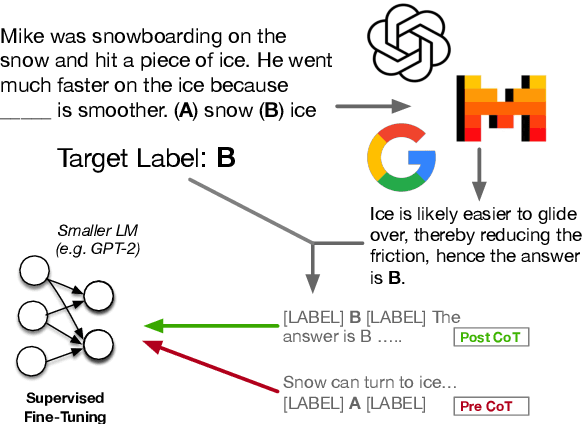
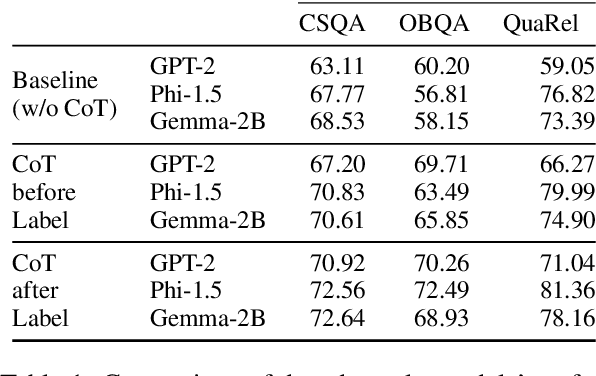
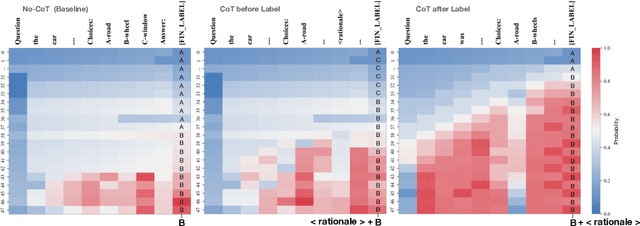
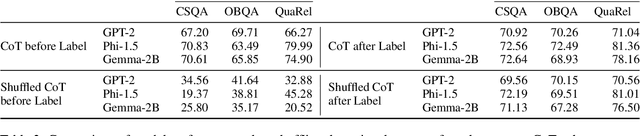
Abstract:Eliciting "chain of thought" (CoT) rationales -- sequences of token that convey a "reasoning" process -- has been shown to consistently improve LLM performance on tasks like question answering. More recent efforts have shown that such rationales can also be used for model distillation: Including CoT sequences (elicited from a large "teacher" model) in addition to target labels when fine-tuning a small student model yields (often substantial) improvements. In this work we ask: Why and how does this additional training signal help in model distillation? We perform ablations to interrogate this, and report some potentially surprising results. Specifically: (1) Placing CoT sequences after labels (rather than before) realizes consistently better downstream performance -- this means that no student "reasoning" is necessary at test time to realize gains. (2) When rationales are appended in this way, they need not be coherent reasoning sequences to yield improvements; performance increases are robust to permutations of CoT tokens, for example. In fact, (3) a small number of key tokens are sufficient to achieve improvements equivalent to those observed when full rationales are used in model distillation.
On-the-fly Definition Augmentation of LLMs for Biomedical NER
Mar 29, 2024Abstract:Despite their general capabilities, LLMs still struggle on biomedical NER tasks, which are difficult due to the presence of specialized terminology and lack of training data. In this work we set out to improve LLM performance on biomedical NER in limited data settings via a new knowledge augmentation approach which incorporates definitions of relevant concepts on-the-fly. During this process, to provide a test bed for knowledge augmentation, we perform a comprehensive exploration of prompting strategies. Our experiments show that definition augmentation is useful for both open source and closed LLMs. For example, it leads to a relative improvement of 15\% (on average) in GPT-4 performance (F1) across all (six) of our test datasets. We conduct extensive ablations and analyses to demonstrate that our performance improvements stem from adding relevant definitional knowledge. We find that careful prompting strategies also improve LLM performance, allowing them to outperform fine-tuned language models in few-shot settings. To facilitate future research in this direction, we release our code at https://github.com/allenai/beacon.
Retrieving Evidence from EHRs with LLMs: Possibilities and Challenges
Sep 08, 2023



Abstract:Unstructured Electronic Health Record (EHR) data often contains critical information complementary to imaging data that would inform radiologists' diagnoses. However, time constraints and the large volume of notes frequently associated with individual patients renders manual perusal of such data to identify relevant evidence infeasible in practice. Modern Large Language Models (LLMs) provide a flexible means of interacting with unstructured EHR data, and may provide a mechanism to efficiently retrieve and summarize unstructured evidence relevant to a given query. In this work, we propose and evaluate an LLM (Flan-T5 XXL) for this purpose. Specifically, in a zero-shot setting we task the LLM to infer whether a patient has or is at risk of a particular condition; if so, we prompt the model to summarize the supporting evidence. Enlisting radiologists for manual evaluation, we find that this LLM-based approach provides outputs consistently preferred to a standard information retrieval baseline, but we also highlight the key outstanding challenge: LLMs are prone to hallucinating evidence. However, we provide results indicating that model confidence in outputs might indicate when LLMs are hallucinating, potentially providing a means to address this.
Revisiting Relation Extraction in the era of Large Language Models
May 08, 2023Abstract:Relation extraction (RE) is the core NLP task of inferring semantic relationships between entities from text. Standard supervised RE techniques entail training modules to tag tokens comprising entity spans and then predict the relationship between them. Recent work has instead treated the problem as a \emph{sequence-to-sequence} task, linearizing relations between entities as target strings to be generated conditioned on the input. Here we push the limits of this approach, using larger language models (GPT-3 and Flan-T5 large) than considered in prior work and evaluating their performance on standard RE tasks under varying levels of supervision. We address issues inherent to evaluating generative approaches to RE by doing human evaluations, in lieu of relying on exact matching. Under this refined evaluation, we find that: (1) Few-shot prompting with GPT-3 achieves near SOTA performance, i.e., roughly equivalent to existing fully supervised models; (2) Flan-T5 is not as capable in the few-shot setting, but supervising and fine-tuning it with Chain-of-Thought (CoT) style explanations (generated via GPT-3) yields SOTA results. We release this model as a new baseline for RE tasks.
Jointly Extracting Interventions, Outcomes, and Findings from RCT Reports with LLMs
May 05, 2023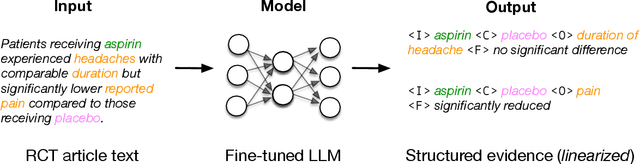
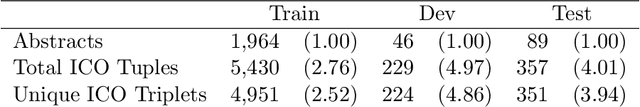
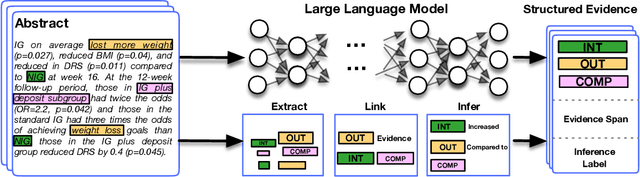
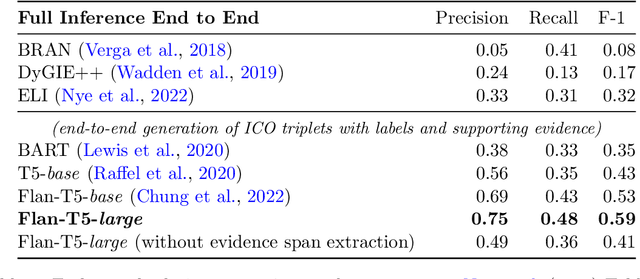
Abstract:Results from Randomized Controlled Trials (RCTs) establish the comparative effectiveness of interventions, and are in turn critical inputs for evidence-based care. However, results from RCTs are presented in (often unstructured) natural language articles describing the design, execution, and outcomes of trials; clinicians must manually extract findings pertaining to interventions and outcomes of interest from such articles. This onerous manual process has motivated work on (semi-)automating extraction of structured evidence from trial reports. In this work we propose and evaluate a text-to-text model built on instruction-tuned Large Language Models (LLMs) to jointly extract Interventions, Outcomes, and Comparators (ICO elements) from clinical abstracts, and infer the associated results reported. Manual (expert) and automated evaluations indicate that framing evidence extraction as a conditional generation task and fine-tuning LLMs for this purpose realizes considerable ($\sim$20 point absolute F1 score) gains over the previous SOTA. We perform ablations and error analyses to assess aspects that contribute to model performance, and to highlight potential directions for further improvements. We apply our model to a collection of published RCTs through mid-2022, and release a searchable database of structured findings (anonymously for now): bit.ly/joint-relations-extraction-mlhc
RedHOT: A Corpus of Annotated Medical Questions, Experiences, and Claims on Social Media
Oct 12, 2022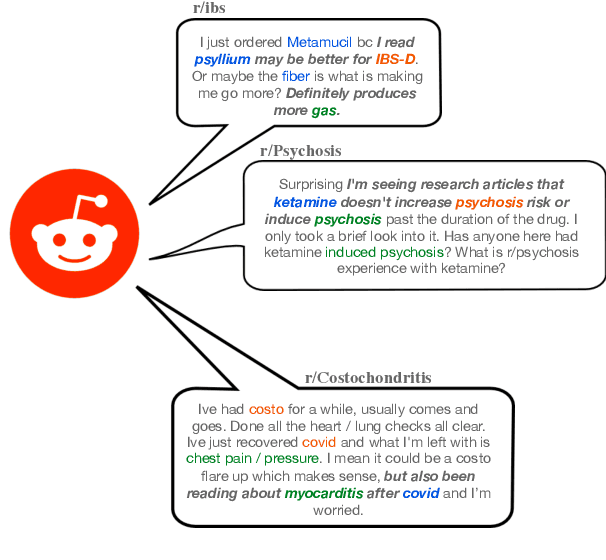
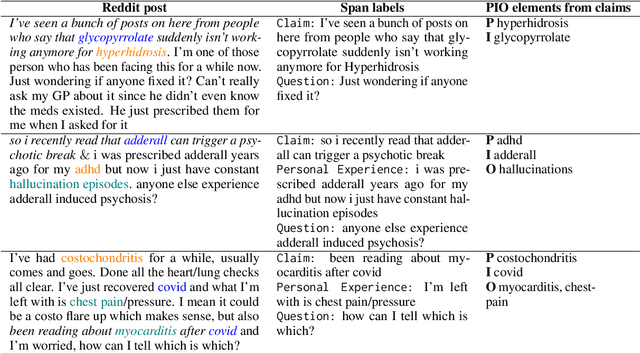
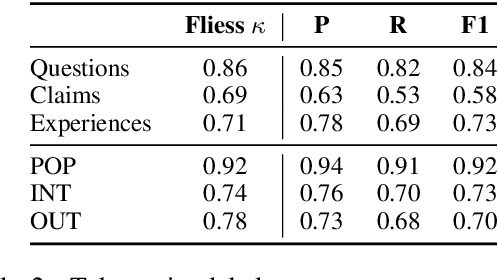
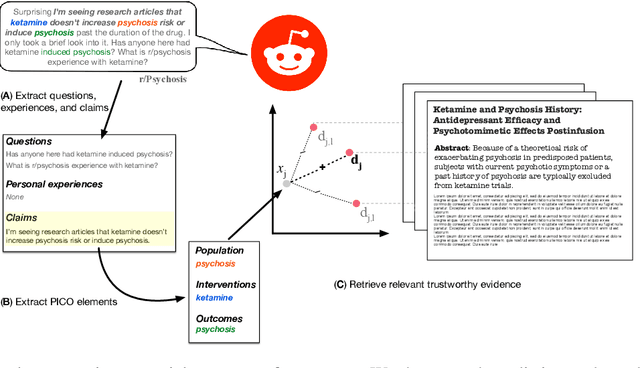
Abstract:We present Reddit Health Online Talk (RedHOT), a corpus of 22,000 richly annotated social media posts from Reddit spanning 24 health conditions. Annotations include demarcations of spans corresponding to medical claims, personal experiences, and questions. We collect additional granular annotations on identified claims. Specifically, we mark snippets that describe patient Populations, Interventions, and Outcomes (PIO elements) within these. Using this corpus, we introduce the task of retrieving trustworthy evidence relevant to a given claim made on social media. We propose a new method to automatically derive (noisy) supervision for this task which we use to train a dense retrieval model; this outperforms baseline models. Manual evaluation of retrieval results performed by medical doctors indicate that while our system performance is promising, there is considerable room for improvement. Collected annotations (and scripts to assemble the dataset), are available at https://github.com/sominw/redhot.
On the Impact of Random Seeds on the Fairness of Clinical Classifiers
Apr 13, 2021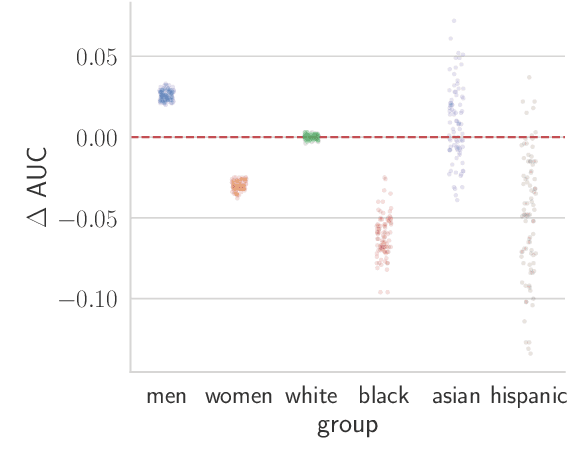
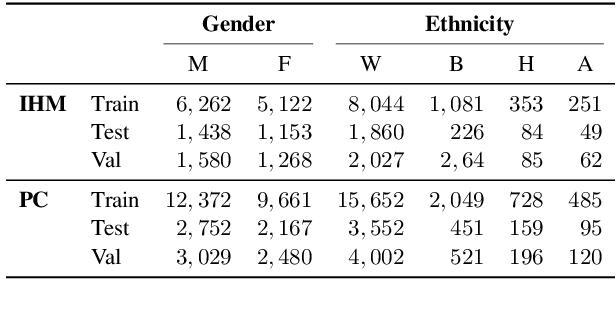
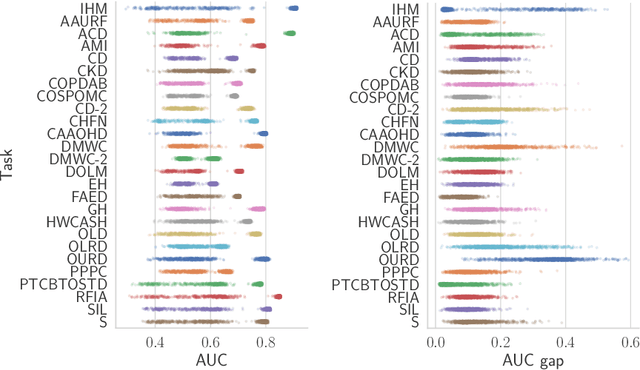
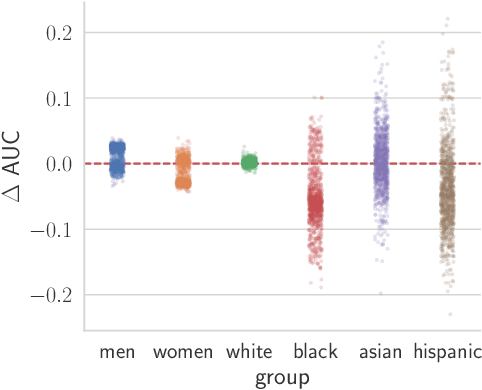
Abstract:Recent work has shown that fine-tuning large networks is surprisingly sensitive to changes in random seed(s). We explore the implications of this phenomenon for model fairness across demographic groups in clinical prediction tasks over electronic health records (EHR) in MIMIC-III -- the standard dataset in clinical NLP research. Apparent subgroup performance varies substantially for seeds that yield similar overall performance, although there is no evidence of a trade-off between overall and subgroup performance. However, we also find that the small sample sizes inherent to looking at intersections of minority groups and somewhat rare conditions limit our ability to accurately estimate disparities. Further, we find that jointly optimizing for high overall performance and low disparities does not yield statistically significant improvements. Our results suggest that fairness work using MIMIC-III should carefully account for variations in apparent differences that may arise from stochasticity and small sample sizes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge