Somin Wadhwa
Who Taught You That? Tracing Teachers in Model Distillation
Feb 10, 2025Abstract:Model distillation -- using outputs from a large teacher model to teach a small student model -- is a practical means of creating efficient models for a particular task. We ask: Can we identify a students' teacher based on its outputs? Such "footprints" left by teacher LLMs would be interesting artifacts. Beyond this, reliable teacher inference may have practical implications as actors seek to distill specific capabilities of massive proprietary LLMs into deployed smaller LMs, potentially violating terms of service. We consider practical task distillation targets including summarization, question answering, and instruction-following. We assume a finite set of candidate teacher models, which we treat as blackboxes. We design discriminative models that operate over lexical features. We find that $n$-gram similarity alone is unreliable for identifying teachers, but part-of-speech (PoS) templates preferred by student models mimic those of their teachers.
Investigating Mysteries of CoT-Augmented Distillation
Jun 20, 2024
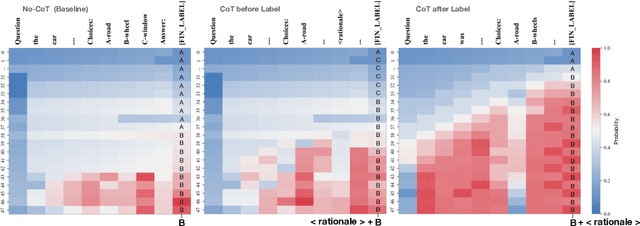
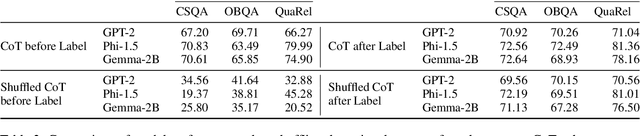
Abstract:Eliciting "chain of thought" (CoT) rationales -- sequences of token that convey a "reasoning" process -- has been shown to consistently improve LLM performance on tasks like question answering. More recent efforts have shown that such rationales can also be used for model distillation: Including CoT sequences (elicited from a large "teacher" model) in addition to target labels when fine-tuning a small student model yields (often substantial) improvements. In this work we ask: Why and how does this additional training signal help in model distillation? We perform ablations to interrogate this, and report some potentially surprising results. Specifically: (1) Placing CoT sequences after labels (rather than before) realizes consistently better downstream performance -- this means that no student "reasoning" is necessary at test time to realize gains. (2) When rationales are appended in this way, they need not be coherent reasoning sequences to yield improvements; performance increases are robust to permutations of CoT tokens, for example. In fact, (3) a small number of key tokens are sufficient to achieve improvements equivalent to those observed when full rationales are used in model distillation.
Learning from Natural Language Explanations for Generalizable Entity Matching
Jun 13, 2024Abstract:Entity matching is the task of linking records from different sources that refer to the same real-world entity. Past work has primarily treated entity linking as a standard supervised learning problem. However, supervised entity matching models often do not generalize well to new data, and collecting exhaustive labeled training data is often cost prohibitive. Further, recent efforts have adopted LLMs for this task in few/zero-shot settings, exploiting their general knowledge. But LLMs are prohibitively expensive for performing inference at scale for real-world entity matching tasks. As an efficient alternative, we re-cast entity matching as a conditional generation task as opposed to binary classification. This enables us to "distill" LLM reasoning into smaller entity matching models via natural language explanations. This approach achieves strong performance, especially on out-of-domain generalization tests (10.85% F-1) where standalone generative methods struggle. We perform ablations that highlight the importance of explanations, both for performance and model robustness.
Distilling Event Sequence Knowledge From Large Language Models
Jan 14, 2024



Abstract:Event sequence models have been found to be highly effective in the analysis and prediction of events. Building such models requires availability of abundant high-quality event sequence data. In certain applications, however, clean structured event sequences are not available, and automated sequence extraction results in data that is too noisy and incomplete. In this work, we explore the use of Large Language Models (LLMs) to generate event sequences that can effectively be used for probabilistic event model construction. This can be viewed as a mechanism of distilling event sequence knowledge from LLMs. Our approach relies on a Knowledge Graph (KG) of event concepts with partial causal relations to guide the generative language model for causal event sequence generation. We show that our approach can generate high-quality event sequences, filling a knowledge gap in the input KG. Furthermore, we explore how the generated sequences can be leveraged to discover useful and more complex structured knowledge from pattern mining and probabilistic event models. We release our sequence generation code and evaluation framework, as well as corpus of event sequence data.
Revisiting Relation Extraction in the era of Large Language Models
May 08, 2023Abstract:Relation extraction (RE) is the core NLP task of inferring semantic relationships between entities from text. Standard supervised RE techniques entail training modules to tag tokens comprising entity spans and then predict the relationship between them. Recent work has instead treated the problem as a \emph{sequence-to-sequence} task, linearizing relations between entities as target strings to be generated conditioned on the input. Here we push the limits of this approach, using larger language models (GPT-3 and Flan-T5 large) than considered in prior work and evaluating their performance on standard RE tasks under varying levels of supervision. We address issues inherent to evaluating generative approaches to RE by doing human evaluations, in lieu of relying on exact matching. Under this refined evaluation, we find that: (1) Few-shot prompting with GPT-3 achieves near SOTA performance, i.e., roughly equivalent to existing fully supervised models; (2) Flan-T5 is not as capable in the few-shot setting, but supervising and fine-tuning it with Chain-of-Thought (CoT) style explanations (generated via GPT-3) yields SOTA results. We release this model as a new baseline for RE tasks.
Jointly Extracting Interventions, Outcomes, and Findings from RCT Reports with LLMs
May 05, 2023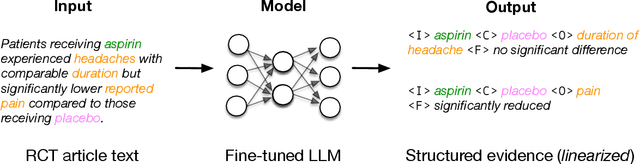
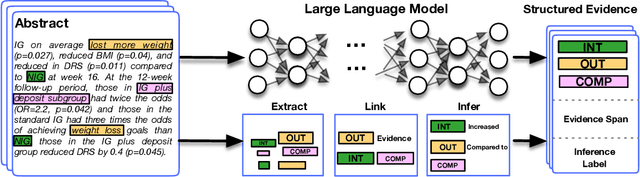
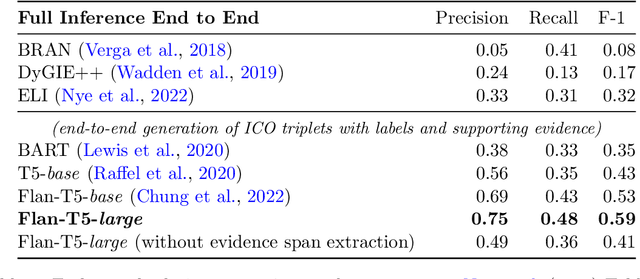
Abstract:Results from Randomized Controlled Trials (RCTs) establish the comparative effectiveness of interventions, and are in turn critical inputs for evidence-based care. However, results from RCTs are presented in (often unstructured) natural language articles describing the design, execution, and outcomes of trials; clinicians must manually extract findings pertaining to interventions and outcomes of interest from such articles. This onerous manual process has motivated work on (semi-)automating extraction of structured evidence from trial reports. In this work we propose and evaluate a text-to-text model built on instruction-tuned Large Language Models (LLMs) to jointly extract Interventions, Outcomes, and Comparators (ICO elements) from clinical abstracts, and infer the associated results reported. Manual (expert) and automated evaluations indicate that framing evidence extraction as a conditional generation task and fine-tuning LLMs for this purpose realizes considerable ($\sim$20 point absolute F1 score) gains over the previous SOTA. We perform ablations and error analyses to assess aspects that contribute to model performance, and to highlight potential directions for further improvements. We apply our model to a collection of published RCTs through mid-2022, and release a searchable database of structured findings (anonymously for now): bit.ly/joint-relations-extraction-mlhc
RedHOT: A Corpus of Annotated Medical Questions, Experiences, and Claims on Social Media
Oct 12, 2022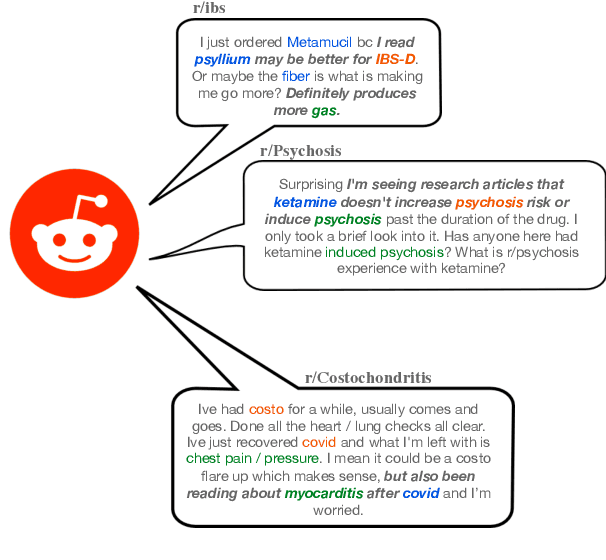
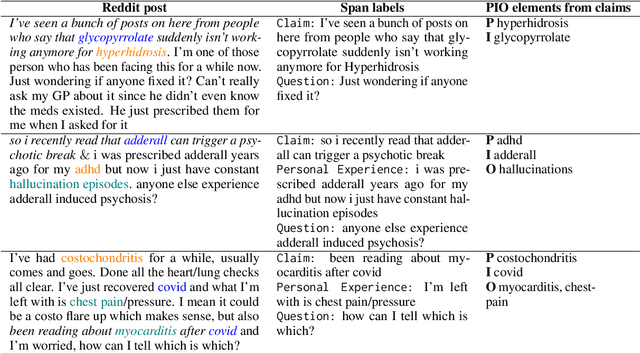
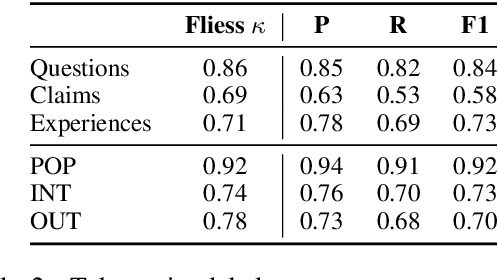
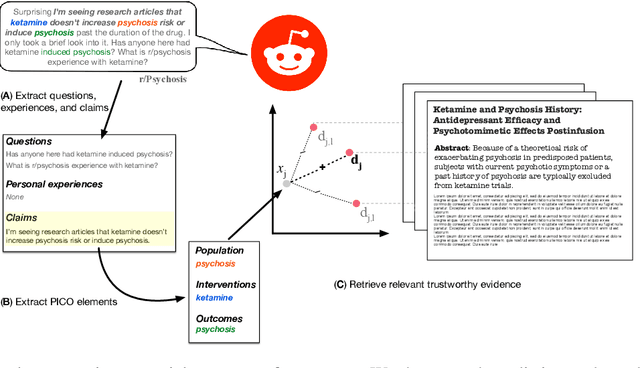
Abstract:We present Reddit Health Online Talk (RedHOT), a corpus of 22,000 richly annotated social media posts from Reddit spanning 24 health conditions. Annotations include demarcations of spans corresponding to medical claims, personal experiences, and questions. We collect additional granular annotations on identified claims. Specifically, we mark snippets that describe patient Populations, Interventions, and Outcomes (PIO elements) within these. Using this corpus, we introduce the task of retrieving trustworthy evidence relevant to a given claim made on social media. We propose a new method to automatically derive (noisy) supervision for this task which we use to train a dense retrieval model; this outperforms baseline models. Manual evaluation of retrieval results performed by medical doctors indicate that while our system performance is promising, there is considerable room for improvement. Collected annotations (and scripts to assemble the dataset), are available at https://github.com/sominw/redhot.
Semi-Automating Knowledge Base Construction for Cancer Genetics
May 26, 2020



Abstract:In this work, we consider the exponentially growing subarea of genetics in cancer. The need to synthesize and centralize this evidence for dissemination has motivated a team of physicians to manually construct and maintain a knowledge base that distills key results reported in the literature. This is a laborious process that entails reading through full-text articles to understand the study design, assess study quality, and extract the reported cancer risk estimates associated with particular hereditary cancer genes (i.e., penetrance). In this work, we propose models to automatically surface key elements from full-text cancer genetics articles, with the ultimate aim of expediting the manual workflow currently in place. We propose two challenging tasks that are critical for characterizing the findings reported cancer genetics studies: (i) Extracting snippets of text that describe \emph{ascertainment mechanisms}, which in turn inform whether the population studied may introduce bias owing to deviations from the target population; (ii) Extracting reported risk estimates (e.g., odds or hazard ratios) associated with specific germline mutations. The latter task may be viewed as a joint entity tagging and relation extraction problem. To train models for these tasks, we induce distant supervision over tokens and snippets in full-text articles using the manually constructed knowledge base. We propose and evaluate several model variants, including a transformer-based joint entity and relation extraction model to extract <germline mutation, risk-estimate>} pairs. We observe strong empirical performance, highlighting the practical potential for such models to aid KB construction in this space. We ablate components of our model, observing, e.g., that a joint model for <germline mutation, risk-estimate> fares substantially better than a pipelined approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge