Sergey Feldman
AstaBench: Rigorous Benchmarking of AI Agents with a Scientific Research Suite
Oct 24, 2025Abstract:AI agents hold the potential to revolutionize scientific productivity by automating literature reviews, replicating experiments, analyzing data, and even proposing new directions of inquiry; indeed, there are now many such agents, ranging from general-purpose "deep research" systems to specialized science-specific agents, such as AI Scientist and AIGS. Rigorous evaluation of these agents is critical for progress. Yet existing benchmarks fall short on several fronts: they (1) fail to provide holistic, product-informed measures of real-world use cases such as science research; (2) lack reproducible agent tools necessary for a controlled comparison of core agentic capabilities; (3) do not account for confounding variables such as model cost and tool access; (4) do not provide standardized interfaces for quick agent prototyping and evaluation; and (5) lack comprehensive baseline agents necessary to identify true advances. In response, we define principles and tooling for more rigorously benchmarking agents. Using these, we present AstaBench, a suite that provides the first holistic measure of agentic ability to perform scientific research, comprising 2400+ problems spanning the entire scientific discovery process and multiple scientific domains, and including many problems inspired by actual user requests to deployed Asta agents. Our suite comes with the first scientific research environment with production-grade search tools that enable controlled, reproducible evaluation, better accounting for confounders. Alongside, we provide a comprehensive suite of nine science-optimized classes of Asta agents and numerous baselines. Our extensive evaluation of 57 agents across 22 agent classes reveals several interesting findings, most importantly that despite meaningful progress on certain individual aspects, AI remains far from solving the challenge of science research assistance.
Ai2 Scholar QA: Organized Literature Synthesis with Attribution
Apr 15, 2025Abstract:Retrieval-augmented generation is increasingly effective in answering scientific questions from literature, but many state-of-the-art systems are expensive and closed-source. We introduce Ai2 Scholar QA, a free online scientific question answering application. To facilitate research, we make our entire pipeline public: as a customizable open-source Python package and interactive web app, along with paper indexes accessible through public APIs and downloadable datasets. We describe our system in detail and present experiments analyzing its key design decisions. In an evaluation on a recent scientific QA benchmark, we find that Ai2 Scholar QA outperforms competing systems.
OpenScholar: Synthesizing Scientific Literature with Retrieval-augmented LMs
Nov 21, 2024



Abstract:Scientific progress depends on researchers' ability to synthesize the growing body of literature. Can large language models (LMs) assist scientists in this task? We introduce OpenScholar, a specialized retrieval-augmented LM that answers scientific queries by identifying relevant passages from 45 million open-access papers and synthesizing citation-backed responses. To evaluate OpenScholar, we develop ScholarQABench, the first large-scale multi-domain benchmark for literature search, comprising 2,967 expert-written queries and 208 long-form answers across computer science, physics, neuroscience, and biomedicine. On ScholarQABench, OpenScholar-8B outperforms GPT-4o by 5% and PaperQA2 by 7% in correctness, despite being a smaller, open model. While GPT4o hallucinates citations 78 to 90% of the time, OpenScholar achieves citation accuracy on par with human experts. OpenScholar's datastore, retriever, and self-feedback inference loop also improves off-the-shelf LMs: for instance, OpenScholar-GPT4o improves GPT-4o's correctness by 12%. In human evaluations, experts preferred OpenScholar-8B and OpenScholar-GPT4o responses over expert-written ones 51% and 70% of the time, respectively, compared to GPT4o's 32%. We open-source all of our code, models, datastore, data and a public demo.
TOPICAL: TOPIC Pages AutomagicaLly
May 03, 2024



Abstract:Topic pages aggregate useful information about an entity or concept into a single succinct and accessible article. Automated creation of topic pages would enable their rapid curation as information resources, providing an alternative to traditional web search. While most prior work has focused on generating topic pages about biographical entities, in this work, we develop a completely automated process to generate high-quality topic pages for scientific entities, with a focus on biomedical concepts. We release TOPICAL, a web app and associated open-source code, comprising a model pipeline combining retrieval, clustering, and prompting, that makes it easy for anyone to generate topic pages for a wide variety of biomedical entities on demand. In a human evaluation of 150 diverse topic pages generated using TOPICAL, we find that the vast majority were considered relevant, accurate, and coherent, with correct supporting citations. We make all code publicly available and host a free-to-use web app at: https://s2-topical.apps.allenai.org
On-the-fly Definition Augmentation of LLMs for Biomedical NER
Mar 29, 2024Abstract:Despite their general capabilities, LLMs still struggle on biomedical NER tasks, which are difficult due to the presence of specialized terminology and lack of training data. In this work we set out to improve LLM performance on biomedical NER in limited data settings via a new knowledge augmentation approach which incorporates definitions of relevant concepts on-the-fly. During this process, to provide a test bed for knowledge augmentation, we perform a comprehensive exploration of prompting strategies. Our experiments show that definition augmentation is useful for both open source and closed LLMs. For example, it leads to a relative improvement of 15\% (on average) in GPT-4 performance (F1) across all (six) of our test datasets. We conduct extensive ablations and analyses to demonstrate that our performance improvements stem from adding relevant definitional knowledge. We find that careful prompting strategies also improve LLM performance, allowing them to outperform fine-tuned language models in few-shot settings. To facilitate future research in this direction, we release our code at https://github.com/allenai/beacon.
RCT Rejection Sampling for Causal Estimation Evaluation
Jul 27, 2023



Abstract:Confounding is a significant obstacle to unbiased estimation of causal effects from observational data. For settings with high-dimensional covariates -- such as text data, genomics, or the behavioral social sciences -- researchers have proposed methods to adjust for confounding by adapting machine learning methods to the goal of causal estimation. However, empirical evaluation of these adjustment methods has been challenging and limited. In this work, we build on a promising empirical evaluation strategy that simplifies evaluation design and uses real data: subsampling randomized controlled trials (RCTs) to create confounded observational datasets while using the average causal effects from the RCTs as ground-truth. We contribute a new sampling algorithm, which we call RCT rejection sampling, and provide theoretical guarantees that causal identification holds in the observational data to allow for valid comparisons to the ground-truth RCT. Using synthetic data, we show our algorithm indeed results in low bias when oracle estimators are evaluated on the confounded samples, which is not always the case for a previously proposed algorithm. In addition to this identification result, we highlight several finite data considerations for evaluation designers who plan to use RCT rejection sampling on their own datasets. As a proof of concept, we implement an example evaluation pipeline and walk through these finite data considerations with a novel, real-world RCT -- which we release publicly -- consisting of approximately 70k observations and text data as high-dimensional covariates. Together, these contributions build towards a broader agenda of improved empirical evaluation for causal estimation.
S2abEL: A Dataset for Entity Linking from Scientific Tables
Apr 30, 2023



Abstract:Entity linking (EL) is the task of linking a textual mention to its corresponding entry in a knowledge base, and is critical for many knowledge-intensive NLP applications. When applied to tables in scientific papers, EL is a step toward large-scale scientific knowledge bases that could enable advanced scientific question answering and analytics. We present the first dataset for EL in scientific tables. EL for scientific tables is especially challenging because scientific knowledge bases can be very incomplete, and disambiguating table mentions typically requires understanding the papers's tet in addition to the table. Our dataset, S2abEL, focuses on EL in machine learning results tables and includes hand-labeled cell types, attributed sources, and entity links from the PaperswithCode taxonomy for 8,429 cells from 732 tables. We introduce a neural baseline method designed for EL on scientific tables containing many out-of-knowledge-base mentions, and show that it significantly outperforms a state-of-the-art generic table EL method. The best baselines fall below human performance, and our analysis highlights avenues for improvement.
The Semantic Scholar Open Data Platform
Jan 24, 2023



Abstract:The volume of scientific output is creating an urgent need for automated tools to help scientists keep up with developments in their field. Semantic Scholar (S2) is an open data platform and website aimed at accelerating science by helping scholars discover and understand scientific literature. We combine public and proprietary data sources using state-of-the-art techniques for scholarly PDF content extraction and automatic knowledge graph construction to build the Semantic Scholar Academic Graph, the largest open scientific literature graph to-date, with 200M+ papers, 80M+ authors, 550M+ paper-authorship edges, and 2.4B+ citation edges. The graph includes advanced semantic features such as structurally parsed text, natural language summaries, and vector embeddings. In this paper, we describe the components of the S2 data processing pipeline and the associated APIs offered by the platform. We will update this living document to reflect changes as we add new data offerings and improve existing services.
SciRepEval: A Multi-Format Benchmark for Scientific Document Representations
Nov 23, 2022



Abstract:Learned representations of scientific documents can serve as valuable input features for downstream tasks, without the need for further fine-tuning. However, existing benchmarks for evaluating these representations fail to capture the diversity of relevant tasks. In response, we introduce SciRepEval, the first comprehensive benchmark for training and evaluating scientific document representations. It includes 25 challenging and realistic tasks, 11 of which are new, across four formats: classification, regression, ranking and search. We then use the benchmark to study and improve the generalization ability of scientific document representation models. We show how state-of-the-art models struggle to generalize across task formats, and that simple multi-task training fails to improve them. However, a new approach that learns multiple embeddings per document, each tailored to a different format, can improve performance. We experiment with task-format-specific control codes and adapters in a multi-task setting and find that they outperform the existing single-embedding state-of-the-art by up to 1.5 points absolute.
Literature-Augmented Clinical Outcome Prediction
Nov 16, 2021

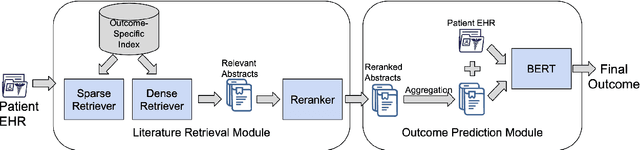

Abstract:Predictive models for medical outcomes hold great promise for enhancing clinical decision-making. These models are trained on rich patient data such as clinical notes, aggregating many patient signals into an outcome prediction. However, AI-based clinical models have typically been developed in isolation from the prominent paradigm of Evidence Based Medicine (EBM), in which medical decisions are based on explicit evidence from existing literature. In this work, we introduce techniques to help bridge this gap between EBM and AI-based clinical models, and show that these methods can improve predictive accuracy. We propose a novel system that automatically retrieves patient-specific literature based on intensive care (ICU) patient information, aggregates relevant papers and fuses them with internal admission notes to form outcome predictions. Our model is able to substantially boost predictive accuracy on three challenging tasks in comparison to strong recent baselines; for in-hospital mortality, we are able to boost top-10% precision by a large margin of over 25%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge