Doug Downey
Allen Institute for Artificial Intelligence, Northwestern University
Generating Literature-Driven Scientific Theories at Scale
Jan 22, 2026Abstract:Contemporary automated scientific discovery has focused on agents for generating scientific experiments, while systems that perform higher-level scientific activities such as theory building remain underexplored. In this work, we formulate the problem of synthesizing theories consisting of qualitative and quantitative laws from large corpora of scientific literature. We study theory generation at scale, using 13.7k source papers to synthesize 2.9k theories, examining how generation using literature-grounding versus parametric knowledge, and accuracy-focused versus novelty-focused generation objectives change theory properties. Our experiments show that, compared to using parametric LLM memory for generation, our literature-supported method creates theories that are significantly better at both matching existing evidence and at predicting future results from 4.6k subsequently-written papers
Preference Learning from Physics-Based Feedback: Tuning Language Models to Design BCC/B2 Superalloys
Nov 15, 2025Abstract:We apply preference learning to the task of language model-guided design of novel structural alloys. In contrast to prior work that focuses on generating stable inorganic crystals, our approach targets the synthesizeability of a specific structural class: BCC/B2 superalloys, an underexplored family of materials with potential applications in extreme environments. Using three open-weight models (LLaMA-3.1, Gemma-2, and OLMo-2), we demonstrate that language models can be optimized for multiple design objectives using a single, unified reward signal through Direct Preference Optimization (DPO). Unlike prior approaches that rely on heuristic or human-in-the-loop feedback (costly), our reward signal is derived from thermodynamic phase calculations, offering a scientifically grounded criterion for model tuning. To our knowledge, this is the first demonstration of preference-tuning a language model using physics-grounded feedback for structural alloy design. The resulting framework is general and extensible, providing a path forward for intelligent design-space exploration across a range of physical science domains.
AstaBench: Rigorous Benchmarking of AI Agents with a Scientific Research Suite
Oct 24, 2025Abstract:AI agents hold the potential to revolutionize scientific productivity by automating literature reviews, replicating experiments, analyzing data, and even proposing new directions of inquiry; indeed, there are now many such agents, ranging from general-purpose "deep research" systems to specialized science-specific agents, such as AI Scientist and AIGS. Rigorous evaluation of these agents is critical for progress. Yet existing benchmarks fall short on several fronts: they (1) fail to provide holistic, product-informed measures of real-world use cases such as science research; (2) lack reproducible agent tools necessary for a controlled comparison of core agentic capabilities; (3) do not account for confounding variables such as model cost and tool access; (4) do not provide standardized interfaces for quick agent prototyping and evaluation; and (5) lack comprehensive baseline agents necessary to identify true advances. In response, we define principles and tooling for more rigorously benchmarking agents. Using these, we present AstaBench, a suite that provides the first holistic measure of agentic ability to perform scientific research, comprising 2400+ problems spanning the entire scientific discovery process and multiple scientific domains, and including many problems inspired by actual user requests to deployed Asta agents. Our suite comes with the first scientific research environment with production-grade search tools that enable controlled, reproducible evaluation, better accounting for confounders. Alongside, we provide a comprehensive suite of nine science-optimized classes of Asta agents and numerous baselines. Our extensive evaluation of 57 agents across 22 agent classes reveals several interesting findings, most importantly that despite meaningful progress on certain individual aspects, AI remains far from solving the challenge of science research assistance.
Demystifying Scientific Problem-Solving in LLMs by Probing Knowledge and Reasoning
Aug 26, 2025Abstract:Scientific problem solving poses unique challenges for LLMs, requiring both deep domain knowledge and the ability to apply such knowledge through complex reasoning. While automated scientific reasoners hold great promise for assisting human scientists, there is currently no widely adopted holistic benchmark for evaluating scientific reasoning, and few approaches systematically disentangle the distinct roles of knowledge and reasoning in these tasks. To address these gaps, we introduce SciReas, a diverse suite of existing benchmarks for scientific reasoning tasks, and SciReas-Pro, a selective subset that requires more complex reasoning. Our holistic evaluation surfaces insights about scientific reasoning performance that remain hidden when relying on individual benchmarks alone. We then propose KRUX, a probing framework for studying the distinct roles of reasoning and knowledge in scientific tasks. Combining the two, we conduct an in-depth analysis that yields several key findings: (1) Retrieving task-relevant knowledge from model parameters is a critical bottleneck for LLMs in scientific reasoning; (2) Reasoning models consistently benefit from external knowledge added in-context on top of the reasoning enhancement; (3) Enhancing verbalized reasoning improves LLMs' ability to surface task-relevant knowledge. Finally, we conduct a lightweight analysis, comparing our science-focused data composition with concurrent efforts on long CoT SFT, and release SciLit01, a strong 8B baseline for scientific reasoning.
SciArena: An Open Evaluation Platform for Foundation Models in Scientific Literature Tasks
Jul 01, 2025
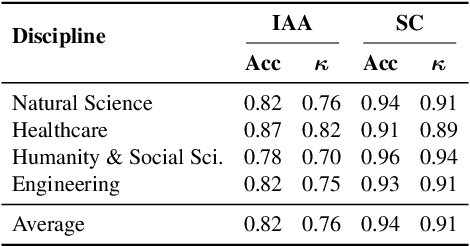

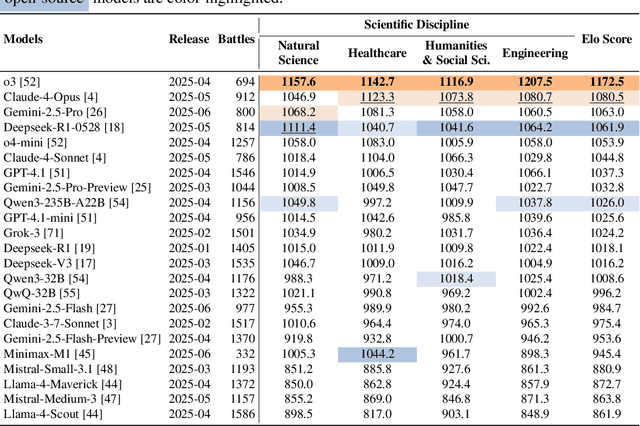
Abstract:We present SciArena, an open and collaborative platform for evaluating foundation models on scientific literature tasks. Unlike traditional benchmarks for scientific literature understanding and synthesis, SciArena engages the research community directly, following the Chatbot Arena evaluation approach of community voting on model comparisons. By leveraging collective intelligence, SciArena offers a community-driven evaluation of model performance on open-ended scientific tasks that demand literature-grounded, long-form responses. The platform currently supports 23 open-source and proprietary foundation models and has collected over 13,000 votes from trusted researchers across diverse scientific domains. We analyze the data collected so far and confirm that the submitted questions are diverse, aligned with real-world literature needs, and that participating researchers demonstrate strong self-consistency and inter-annotator agreement in their evaluations. We discuss the results and insights based on the model ranking leaderboard. To further promote research in building model-based automated evaluation systems for literature tasks, we release SciArena-Eval, a meta-evaluation benchmark based on our collected preference data. The benchmark measures the accuracy of models in judging answer quality by comparing their pairwise assessments with human votes. Our experiments highlight the benchmark's challenges and emphasize the need for more reliable automated evaluation methods.
Ai2 Scholar QA: Organized Literature Synthesis with Attribution
Apr 15, 2025Abstract:Retrieval-augmented generation is increasingly effective in answering scientific questions from literature, but many state-of-the-art systems are expensive and closed-source. We introduce Ai2 Scholar QA, a free online scientific question answering application. To facilitate research, we make our entire pipeline public: as a customizable open-source Python package and interactive web app, along with paper indexes accessible through public APIs and downloadable datasets. We describe our system in detail and present experiments analyzing its key design decisions. In an evaluation on a recent scientific QA benchmark, we find that Ai2 Scholar QA outperforms competing systems.
OpenScholar: Synthesizing Scientific Literature with Retrieval-augmented LMs
Nov 21, 2024



Abstract:Scientific progress depends on researchers' ability to synthesize the growing body of literature. Can large language models (LMs) assist scientists in this task? We introduce OpenScholar, a specialized retrieval-augmented LM that answers scientific queries by identifying relevant passages from 45 million open-access papers and synthesizing citation-backed responses. To evaluate OpenScholar, we develop ScholarQABench, the first large-scale multi-domain benchmark for literature search, comprising 2,967 expert-written queries and 208 long-form answers across computer science, physics, neuroscience, and biomedicine. On ScholarQABench, OpenScholar-8B outperforms GPT-4o by 5% and PaperQA2 by 7% in correctness, despite being a smaller, open model. While GPT4o hallucinates citations 78 to 90% of the time, OpenScholar achieves citation accuracy on par with human experts. OpenScholar's datastore, retriever, and self-feedback inference loop also improves off-the-shelf LMs: for instance, OpenScholar-GPT4o improves GPT-4o's correctness by 12%. In human evaluations, experts preferred OpenScholar-8B and OpenScholar-GPT4o responses over expert-written ones 51% and 70% of the time, respectively, compared to GPT4o's 32%. We open-source all of our code, models, datastore, data and a public demo.
SciRIFF: A Resource to Enhance Language Model Instruction-Following over Scientific Literature
Jun 10, 2024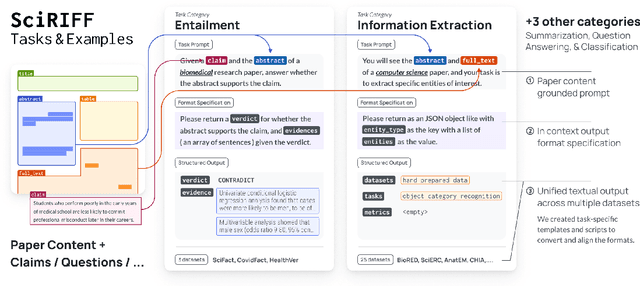
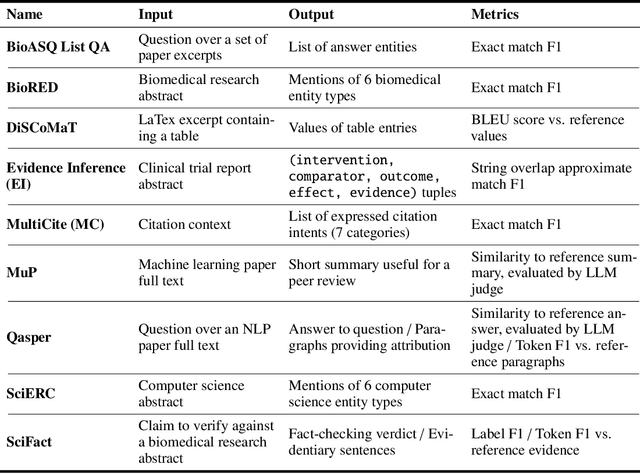
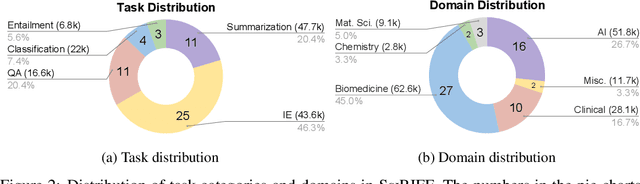
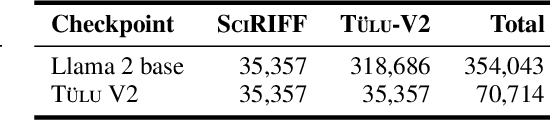
Abstract:We present SciRIFF (Scientific Resource for Instruction-Following and Finetuning), a dataset of 137K instruction-following demonstrations for 54 tasks covering five essential scientific literature understanding capabilities: information extraction, summarization, question answering, claim verification, and classification. SciRIFF demonstrations are notable for their long input contexts, detailed task specifications, and complex structured outputs. While instruction-following resources are available in specific domains such as clinical medicine and chemistry, SciRIFF is the first dataset focused on extracting and synthesizing information from research literature across a wide range of scientific fields. To demonstrate the utility of SciRIFF, we develop a sample-efficient strategy to adapt a general instruction-following model for science by performing additional finetuning on a mix of general-domain and SciRIFF demonstrations. In evaluations on nine held-out scientific tasks, our model -- called SciTulu -- improves over a strong LLM baseline by 28.1% and 6.5% at the 7B and 70B scales respectively, while maintaining general instruction-following performance within 2% of the baseline. We are optimistic that SciRIFF will facilitate the development and evaluation of LLMs to help researchers navigate the ever-growing body of scientific literature. We release our dataset, model checkpoints, and data processing and evaluation code to enable further research.
TOPICAL: TOPIC Pages AutomagicaLly
May 03, 2024



Abstract:Topic pages aggregate useful information about an entity or concept into a single succinct and accessible article. Automated creation of topic pages would enable their rapid curation as information resources, providing an alternative to traditional web search. While most prior work has focused on generating topic pages about biographical entities, in this work, we develop a completely automated process to generate high-quality topic pages for scientific entities, with a focus on biomedical concepts. We release TOPICAL, a web app and associated open-source code, comprising a model pipeline combining retrieval, clustering, and prompting, that makes it easy for anyone to generate topic pages for a wide variety of biomedical entities on demand. In a human evaluation of 150 diverse topic pages generated using TOPICAL, we find that the vast majority were considered relevant, accurate, and coherent, with correct supporting citations. We make all code publicly available and host a free-to-use web app at: https://s2-topical.apps.allenai.org
MARG: Multi-Agent Review Generation for Scientific Papers
Jan 08, 2024Abstract:We study the ability of LLMs to generate feedback for scientific papers and develop MARG, a feedback generation approach using multiple LLM instances that engage in internal discussion. By distributing paper text across agents, MARG can consume the full text of papers beyond the input length limitations of the base LLM, and by specializing agents and incorporating sub-tasks tailored to different comment types (experiments, clarity, impact) it improves the helpfulness and specificity of feedback. In a user study, baseline methods using GPT-4 were rated as producing generic or very generic comments more than half the time, and only 1.7 comments per paper were rated as good overall in the best baseline. Our system substantially improves the ability of GPT-4 to generate specific and helpful feedback, reducing the rate of generic comments from 60% to 29% and generating 3.7 good comments per paper (a 2.2x improvement).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge