Kaiyan Zhang
MARTI-MARS$^2$: Scaling Multi-Agent Self-Search via Reinforcement Learning for Code Generation
Feb 08, 2026Abstract:While the complex reasoning capability of Large Language Models (LLMs) has attracted significant attention, single-agent systems often encounter inherent performance ceilings in complex tasks such as code generation. Multi-agent collaboration offers a promising avenue to transcend these boundaries. However, existing frameworks typically rely on prompt-based test-time interactions or multi-role configurations trained with homogeneous parameters, limiting error correction capabilities and strategic diversity. In this paper, we propose a Multi-Agent Reinforced Training and Inference Framework with Self-Search Scaling (MARTI-MARS2), which integrates policy learning with multi-agent tree search by formulating the multi-agent collaborative exploration process as a dynamic and learnable environment. By allowing agents to iteratively explore and refine within the environment, the framework facilitates evolution from parameter-sharing homogeneous multi-role training to heterogeneous multi-agent training, breaking through single-agent capability limits. We also introduce an efficient inference strategy MARTI-MARS2-T+ to fully exploit the scaling potential of multi-agent collaboration at test time. We conduct extensive experiments across varied model scales (8B, 14B, and 32B) on challenging code generation benchmarks. Utilizing two collaborating 32B models, MARTI-MARS2 achieves 77.7%, outperforming strong baselines like GPT-5.1. Furthermore, MARTI-MARS2 reveals a novel scaling law: shifting from single-agent to homogeneous multi-role and ultimately to heterogeneous multi-agent paradigms progressively yields higher RL performance ceilings, robust TTS capabilities, and greater policy diversity, suggesting that policy diversity is critical for scaling intelligence via multi-agent reinforcement learning.
Emotion-Director: Bridging Affective Shortcut in Emotion-Oriented Image Generation
Dec 22, 2025Abstract:Image generation based on diffusion models has demonstrated impressive capability, motivating exploration into diverse and specialized applications. Owing to the importance of emotion in advertising, emotion-oriented image generation has attracted increasing attention. However, current emotion-oriented methods suffer from an affective shortcut, where emotions are approximated to semantics. As evidenced by two decades of research, emotion is not equivalent to semantics. To this end, we propose Emotion-Director, a cross-modal collaboration framework consisting of two modules. First, we propose a cross-Modal Collaborative diffusion model, abbreviated as MC-Diffusion. MC-Diffusion integrates visual prompts with textual prompts for guidance, enabling the generation of emotion-oriented images beyond semantics. Further, we improve the DPO optimization by a negative visual prompt, enhancing the model's sensitivity to different emotions under the same semantics. Second, we propose MC-Agent, a cross-Modal Collaborative Agent system that rewrites textual prompts to express the intended emotions. To avoid template-like rewrites, MC-Agent employs multi-agents to simulate human subjectivity toward emotions, and adopts a chain-of-concept workflow that improves the visual expressiveness of the rewritten prompts. Extensive qualitative and quantitative experiments demonstrate the superiority of Emotion-Director in emotion-oriented image generation.
JustRL: Scaling a 1.5B LLM with a Simple RL Recipe
Dec 18, 2025Abstract:Recent advances in reinforcement learning for large language models have converged on increasing complexity: multi-stage training pipelines, dynamic hyperparameter schedules, and curriculum learning strategies. This raises a fundamental question: \textbf{Is this complexity necessary?} We present \textbf{JustRL}, a minimal approach using single-stage training with fixed hyperparameters that achieves state-of-the-art performance on two 1.5B reasoning models (54.9\% and 64.3\% average accuracy across nine mathematical benchmarks) while using 2$\times$ less compute than sophisticated approaches. The same hyperparameters transfer across both models without tuning, and training exhibits smooth, monotonic improvement over 4,000+ steps without the collapses or plateaus that typically motivate interventions. Critically, ablations reveal that adding ``standard tricks'' like explicit length penalties and robust verifiers may degrade performance by collapsing exploration. These results suggest that the field may be adding complexity to solve problems that disappear with a stable, scaled-up baseline. We release our models and code to establish a simple, validated baseline for the community.
Attention as a Compass: Efficient Exploration for Process-Supervised RL in Reasoning Models
Sep 30, 2025Abstract:Reinforcement Learning (RL) has shown remarkable success in enhancing the reasoning capabilities of Large Language Models (LLMs). Process-Supervised RL (PSRL) has emerged as a more effective paradigm compared to outcome-based RL. However, existing PSRL approaches suffer from limited exploration efficiency, both in terms of branching positions and sampling. In this paper, we introduce a novel PSRL framework (AttnRL), which enables efficient exploration for reasoning models. Motivated by preliminary observations that steps exhibiting high attention scores correlate with reasoning behaviors, we propose to branch from positions with high values. Furthermore, we develop an adaptive sampling strategy that accounts for problem difficulty and historical batch size, ensuring that the whole training batch maintains non-zero advantage values. To further improve sampling efficiency, we design a one-step off-policy training pipeline for PSRL. Extensive experiments on multiple challenging mathematical reasoning benchmarks demonstrate that our method consistently outperforms prior approaches in terms of performance and sampling and training efficiency.
FlowRL: Matching Reward Distributions for LLM Reasoning
Sep 18, 2025Abstract:We propose FlowRL: matching the full reward distribution via flow balancing instead of maximizing rewards in large language model (LLM) reinforcement learning (RL). Recent advanced reasoning models adopt reward-maximizing methods (\eg, PPO and GRPO), which tend to over-optimize dominant reward signals while neglecting less frequent but valid reasoning paths, thus reducing diversity. In contrast, we transform scalar rewards into a normalized target distribution using a learnable partition function, and then minimize the reverse KL divergence between the policy and the target distribution. We implement this idea as a flow-balanced optimization method that promotes diverse exploration and generalizable reasoning trajectories. We conduct experiments on math and code reasoning tasks: FlowRL achieves a significant average improvement of $10.0\%$ over GRPO and $5.1\%$ over PPO on math benchmarks, and performs consistently better on code reasoning tasks. These results highlight reward distribution-matching as a key step toward efficient exploration and diverse reasoning in LLM reinforcement learning.
SimpleVLA-RL: Scaling VLA Training via Reinforcement Learning
Sep 11, 2025Abstract:Vision-Language-Action (VLA) models have recently emerged as a powerful paradigm for robotic manipulation. Despite substantial progress enabled by large-scale pretraining and supervised fine-tuning (SFT), these models face two fundamental challenges: (i) the scarcity and high cost of large-scale human-operated robotic trajectories required for SFT scaling, and (ii) limited generalization to tasks involving distribution shift. Recent breakthroughs in Large Reasoning Models (LRMs) demonstrate that reinforcement learning (RL) can dramatically enhance step-by-step reasoning capabilities, raising a natural question: Can RL similarly improve the long-horizon step-by-step action planning of VLA? In this work, we introduce SimpleVLA-RL, an efficient RL framework tailored for VLA models. Building upon veRL, we introduce VLA-specific trajectory sampling, scalable parallelization, multi-environment rendering, and optimized loss computation. When applied to OpenVLA-OFT, SimpleVLA-RL achieves SoTA performance on LIBERO and even outperforms $\pi_0$ on RoboTwin 1.0\&2.0 with the exploration-enhancing strategies we introduce. SimpleVLA-RL not only reduces dependence on large-scale data and enables robust generalization, but also remarkably surpasses SFT in real-world tasks. Moreover, we identify a novel phenomenon ``pushcut'' during RL training, wherein the policy discovers previously unseen patterns beyond those seen in the previous training process. Github: https://github.com/PRIME-RL/SimpleVLA-RL
A Survey of Reinforcement Learning for Large Reasoning Models
Sep 10, 2025Abstract:In this paper, we survey recent advances in Reinforcement Learning (RL) for reasoning with Large Language Models (LLMs). RL has achieved remarkable success in advancing the frontier of LLM capabilities, particularly in addressing complex logical tasks such as mathematics and coding. As a result, RL has emerged as a foundational methodology for transforming LLMs into LRMs. With the rapid progress of the field, further scaling of RL for LRMs now faces foundational challenges not only in computational resources but also in algorithm design, training data, and infrastructure. To this end, it is timely to revisit the development of this domain, reassess its trajectory, and explore strategies to enhance the scalability of RL toward Artificial SuperIntelligence (ASI). In particular, we examine research applying RL to LLMs and LRMs for reasoning abilities, especially since the release of DeepSeek-R1, including foundational components, core problems, training resources, and downstream applications, to identify future opportunities and directions for this rapidly evolving area. We hope this review will promote future research on RL for broader reasoning models. Github: https://github.com/TsinghuaC3I/Awesome-RL-for-LRMs
AdsQA: Towards Advertisement Video Understanding
Sep 10, 2025Abstract:Large language models (LLMs) have taken a great step towards AGI. Meanwhile, an increasing number of domain-specific problems such as math and programming boost these general-purpose models to continuously evolve via learning deeper expertise. Now is thus the time further to extend the diversity of specialized applications for knowledgeable LLMs, though collecting high quality data with unexpected and informative tasks is challenging. In this paper, we propose to use advertisement (ad) videos as a challenging test-bed to probe the ability of LLMs in perceiving beyond the objective physical content of common visual domain. Our motivation is to take full advantage of the clue-rich and information-dense ad videos' traits, e.g., marketing logic, persuasive strategies, and audience engagement. Our contribution is three-fold: (1) To our knowledge, this is the first attempt to use ad videos with well-designed tasks to evaluate LLMs. We contribute AdsQA, a challenging ad Video QA benchmark derived from 1,544 ad videos with 10,962 clips, totaling 22.7 hours, providing 5 challenging tasks. (2) We propose ReAd-R, a Deepseek-R1 styled RL model that reflects on questions, and generates answers via reward-driven optimization. (3) We benchmark 14 top-tier LLMs on AdsQA, and our \texttt{ReAd-R}~achieves the state-of-the-art outperforming strong competitors equipped with long-chain reasoning capabilities by a clear margin.
Towards a Unified View of Large Language Model Post-Training
Sep 04, 2025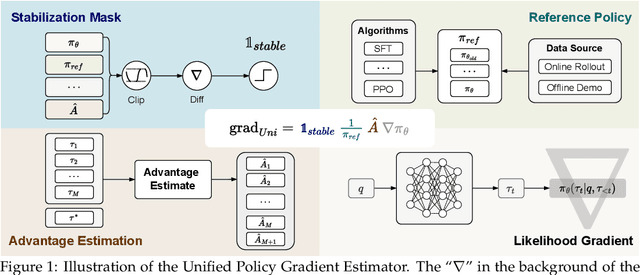
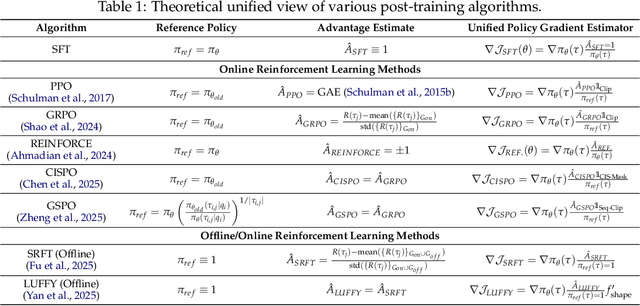
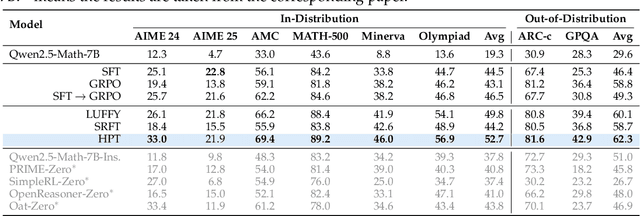
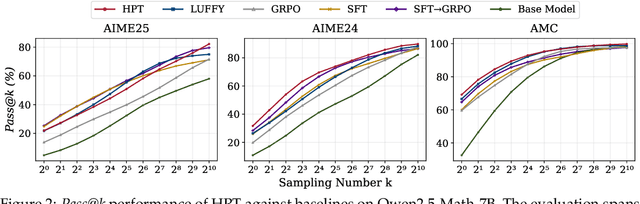
Abstract:Two major sources of training data exist for post-training modern language models: online (model-generated rollouts) data, and offline (human or other-model demonstrations) data. These two types of data are typically used by approaches like Reinforcement Learning (RL) and Supervised Fine-Tuning (SFT), respectively. In this paper, we show that these approaches are not in contradiction, but are instances of a single optimization process. We derive a Unified Policy Gradient Estimator, and present the calculations of a wide spectrum of post-training approaches as the gradient of a common objective under different data distribution assumptions and various bias-variance tradeoffs. The gradient estimator is constructed with four interchangeable parts: stabilization mask, reference policy denominator, advantage estimate, and likelihood gradient. Motivated by our theoretical findings, we propose Hybrid Post-Training (HPT), an algorithm that dynamically selects different training signals. HPT is designed to yield both effective exploitation of demonstration and stable exploration without sacrificing learned reasoning patterns. We provide extensive experiments and ablation studies to verify the effectiveness of our unified theoretical framework and HPT. Across six mathematical reasoning benchmarks and two out-of-distribution suites, HPT consistently surpasses strong baselines across models of varying scales and families.
ReviewRL: Towards Automated Scientific Review with RL
Aug 14, 2025Abstract:Peer review is essential for scientific progress but faces growing challenges due to increasing submission volumes and reviewer fatigue. Existing automated review approaches struggle with factual accuracy, rating consistency, and analytical depth, often generating superficial or generic feedback lacking the insights characteristic of high-quality human reviews. We introduce ReviewRL, a reinforcement learning framework for generating comprehensive and factually grounded scientific paper reviews. Our approach combines: (1) an ArXiv-MCP retrieval-augmented context generation pipeline that incorporates relevant scientific literature, (2) supervised fine-tuning that establishes foundational reviewing capabilities, and (3) a reinforcement learning procedure with a composite reward function that jointly enhances review quality and rating accuracy. Experiments on ICLR 2025 papers demonstrate that ReviewRL significantly outperforms existing methods across both rule-based metrics and model-based quality assessments. ReviewRL establishes a foundational framework for RL-driven automatic critique generation in scientific discovery, demonstrating promising potential for future development in this domain. The implementation of ReviewRL will be released at GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge