Li Sheng
P1: Mastering Physics Olympiads with Reinforcement Learning
Nov 17, 2025Abstract:Recent progress in large language models (LLMs) has moved the frontier from puzzle-solving to science-grade reasoning-the kind needed to tackle problems whose answers must stand against nature, not merely fit a rubric. Physics is the sharpest test of this shift, which binds symbols to reality in a fundamental way, serving as the cornerstone of most modern technologies. In this work, we manage to advance physics research by developing large language models with exceptional physics reasoning capabilities, especially excel at solving Olympiad-level physics problems. We introduce P1, a family of open-source physics reasoning models trained entirely through reinforcement learning (RL). Among them, P1-235B-A22B is the first open-source model with Gold-medal performance at the latest International Physics Olympiad (IPhO 2025), and wins 12 gold medals out of 13 international/regional physics competitions in 2024/2025. P1-30B-A3B also surpasses almost all other open-source models on IPhO 2025, getting a silver medal. Further equipped with an agentic framework PhysicsMinions, P1-235B-A22B+PhysicsMinions achieves overall No.1 on IPhO 2025, and obtains the highest average score over the 13 physics competitions. Besides physics, P1 models also present great performance on other reasoning tasks like math and coding, showing the great generalibility of P1 series.
TTRL: Test-Time Reinforcement Learning
Apr 22, 2025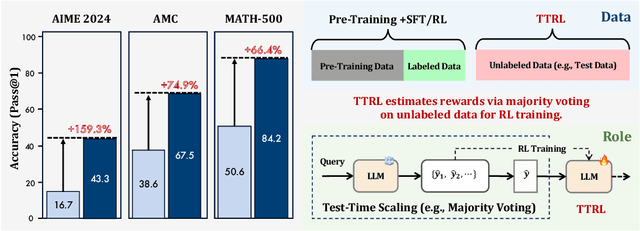
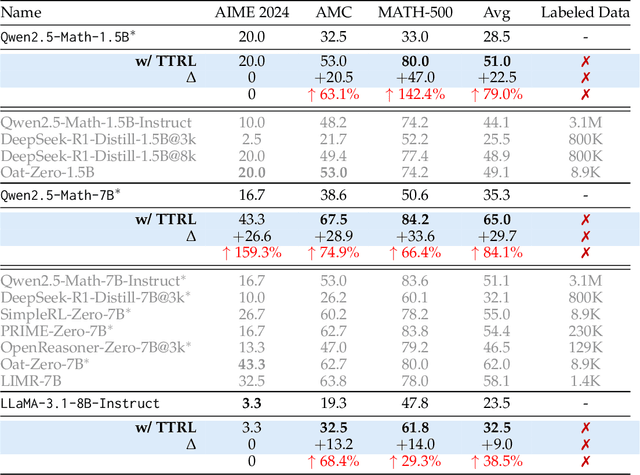
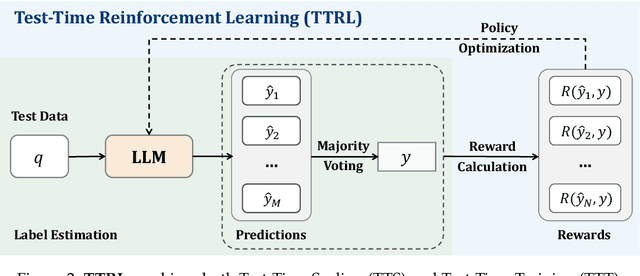
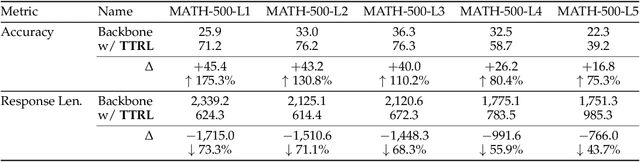
Abstract:This paper investigates Reinforcement Learning (RL) on data without explicit labels for reasoning tasks in Large Language Models (LLMs). The core challenge of the problem is reward estimation during inference while not having access to ground-truth information. While this setting appears elusive, we find that common practices in Test-Time Scaling (TTS), such as majority voting, yield surprisingly effective rewards suitable for driving RL training. In this work, we introduce Test-Time Reinforcement Learning (TTRL), a novel method for training LLMs using RL on unlabeled data. TTRL enables self-evolution of LLMs by utilizing the priors in the pre-trained models. Our experiments demonstrate that TTRL consistently improves performance across a variety of tasks and models. Notably, TTRL boosts the pass@1 performance of Qwen-2.5-Math-7B by approximately 159% on the AIME 2024 with only unlabeled test data. Furthermore, although TTRL is only supervised by the Maj@N metric, TTRL has demonstrated performance to consistently surpass the upper limit of the initial model, and approach the performance of models trained directly on test data with ground-truth labels. Our experimental findings validate the general effectiveness of TTRL across various tasks, and highlight TTRL's potential for broader tasks and domains. GitHub: https://github.com/PRIME-RL/TTRL
UltraIF: Advancing Instruction Following from the Wild
Feb 06, 2025



Abstract:Instruction-following made modern large language models (LLMs) helpful assistants. However, the key to taming LLMs on complex instructions remains mysterious, for that there are huge gaps between models trained by open-source community and those trained by leading companies. To bridge the gap, we propose a simple and scalable approach UltraIF for building LLMs that can follow complex instructions with open-source data. UltraIF first decomposes real-world user prompts into simpler queries, constraints, and corresponding evaluation questions for the constraints. Then, we train an UltraComposer to compose constraint-associated prompts with evaluation questions. This prompt composer allows us to synthesize complicated instructions as well as filter responses with evaluation questions. In our experiment, for the first time, we successfully align LLaMA-3.1-8B-Base to catch up with its instruct version on 5 instruction-following benchmarks without any benchmark information, using only 8B model as response generator and evaluator. The aligned model also achieved competitive scores on other benchmarks. Moreover, we also show that UltraIF could further improve LLaMA-3.1-8B-Instruct through self-alignment, motivating broader use cases for the method. Our code will be available at https://github.com/kkk-an/UltraIF.
Depression Detection on Social Media with Large Language Models
Mar 16, 2024



Abstract:Depression harms. However, due to a lack of mental health awareness and fear of stigma, many patients do not actively seek diagnosis and treatment, leading to detrimental outcomes. Depression detection aims to determine whether an individual suffers from depression by analyzing their history of posts on social media, which can significantly aid in early detection and intervention. It mainly faces two key challenges: 1) it requires professional medical knowledge, and 2) it necessitates both high accuracy and explainability. To address it, we propose a novel depression detection system called DORIS, combining medical knowledge and the recent advances in large language models (LLMs). Specifically, to tackle the first challenge, we proposed an LLM-based solution to first annotate whether high-risk texts meet medical diagnostic criteria. Further, we retrieve texts with high emotional intensity and summarize critical information from the historical mood records of users, so-called mood courses. To tackle the second challenge, we combine LLM and traditional classifiers to integrate medical knowledge-guided features, for which the model can also explain its prediction results, achieving both high accuracy and explainability. Extensive experimental results on benchmarking datasets show that, compared to the current best baseline, our approach improves by 0.036 in AUPRC, which can be considered significant, demonstrating the effectiveness of our approach and its high value as an NLP application.
Identify Critical Nodes in Complex Network with Large Language Models
Mar 01, 2024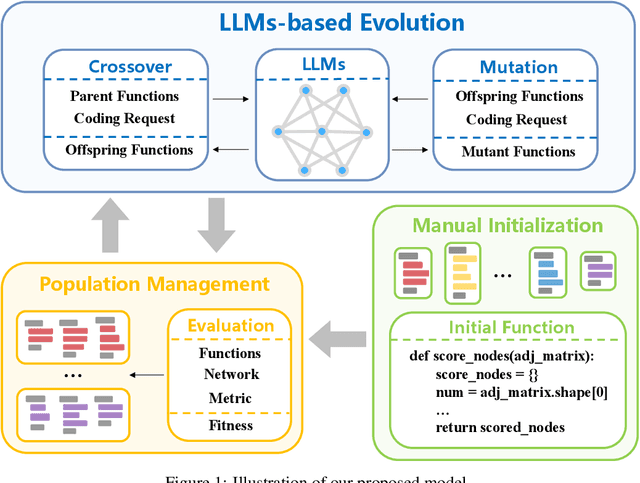
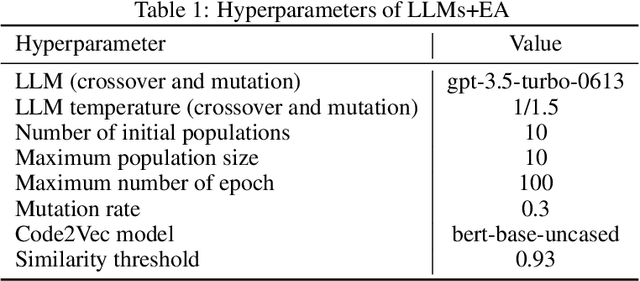
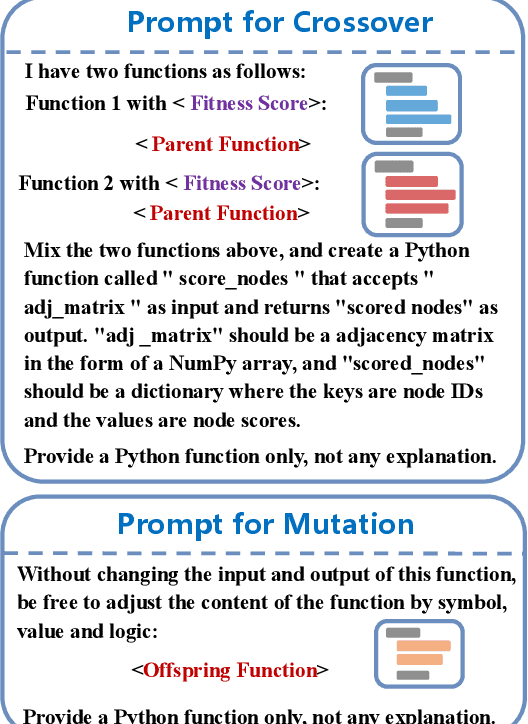
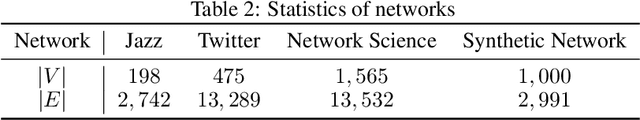
Abstract:Identifying critical nodes in networks is a classical decision-making task, and many methods struggle to strike a balance between adaptability and utility. Therefore, we propose an approach that empowers Evolutionary Algorithm (EA) with Large Language Models (LLMs), to generate a function called "score\_nodes" which can further be used to identify crucial nodes based on their assigned scores. Our model consists of three main components: Manual Initialization, Population Management, and LLMs-based Evolution. It evolves from initial populations with a set of designed node scoring functions created manually. LLMs leverage their strong contextual understanding and rich programming skills to perform crossover and mutation operations on the individuals, generating excellent new functions. These functions are then categorized, ranked, and eliminated to ensure the stable development of the populations while preserving diversity. Extensive experiments demonstrate the excellent performance of our method, showcasing its strong generalization ability compared to other state-of-the-art algorithms. It can consistently and orderly generate diverse and efficient node scoring functions. All source codes and models that can reproduce all results in this work are publicly available at this link: \url{https://anonymous.4open.science/r/LLM4CN-6520}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge