Shenghua He
Facilitating Long Context Understanding via Supervised Chain-of-Thought Reasoning
Feb 18, 2025Abstract:Recent advances in Large Language Models (LLMs) have enabled them to process increasingly longer sequences, ranging from 2K to 2M tokens and even beyond. However, simply extending the input sequence length does not necessarily lead to effective long-context understanding. In this study, we integrate Chain-of-Thought (CoT) reasoning into LLMs in a supervised manner to facilitate effective long-context understanding. To achieve this, we introduce LongFinanceQA, a synthetic dataset in the financial domain designed to improve long-context reasoning. Unlike existing long-context synthetic data, LongFinanceQA includes intermediate CoT reasoning before the final conclusion, which encourages LLMs to perform explicit reasoning, improving accuracy and interpretability in long-context understanding. To generate synthetic CoT reasoning, we propose Property-driven Agentic Inference (PAI), an agentic framework that simulates human-like reasoning steps, including property extraction, retrieval, and summarization. We evaluate PAI's reasoning capabilities by assessing GPT-4o-mini w/ PAI on the Loong benchmark, outperforming standard GPT-4o-mini by 20.0%. Furthermore, we fine-tune LLaMA-3.1-8B-Instruct on LongFinanceQA, achieving a 24.6% gain on Loong's financial subset.
Systematic Evaluation of LLM-as-a-Judge in LLM Alignment Tasks: Explainable Metrics and Diverse Prompt Templates
Aug 23, 2024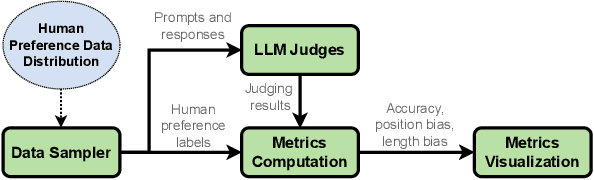
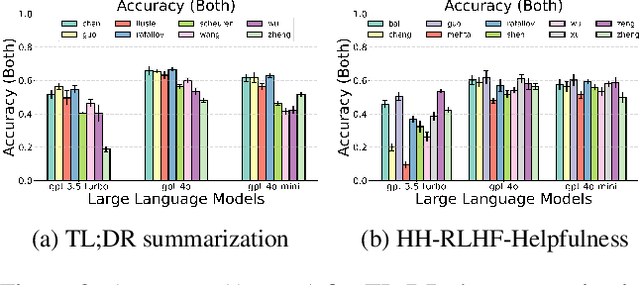
Abstract:Alignment approaches such as RLHF and DPO are actively investigated to align large language models (LLMs) with human preferences. Commercial large language models (LLMs) like GPT-4 have been recently employed to evaluate and compare different LLM alignment approaches. These models act as surrogates for human evaluators due to their promising abilities to approximate human preferences with remarkably faster feedback and lower costs. This methodology is referred to as LLM-as-a-judge. However, concerns regarding its reliability have emerged, attributed to LLM judges' biases and inconsistent decision-making. Previous research has sought to develop robust evaluation frameworks for assessing the reliability of LLM judges and their alignment with human preferences. However, the employed evaluation metrics often lack adequate explainability and fail to address the internal inconsistency of LLMs. Additionally, existing studies inadequately explore the impact of various prompt templates when applying LLM-as-a-judge methods, which leads to potentially inconsistent comparisons between different alignment algorithms. In this work, we systematically evaluate LLM judges on alignment tasks (e.g. summarization) by defining evaluation metrics with improved theoretical interpretability and disentangling reliability metrics with LLM internal inconsistency. We develop a framework to evaluate, compare, and visualize the reliability and alignment of LLM judges to provide informative observations that help choose LLM judges for alignment tasks. Our results indicate a significant impact of prompt templates on LLM judge performance, as well as a mediocre alignment level between the tested LLM judges and human evaluators.
Accurate and Robust Lesion RECIST Diameter Prediction and Segmentation with Transformers
Aug 28, 2022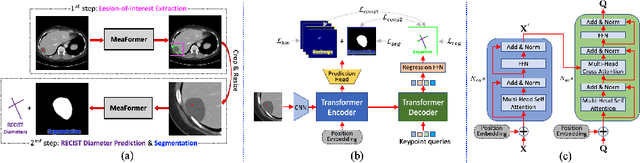
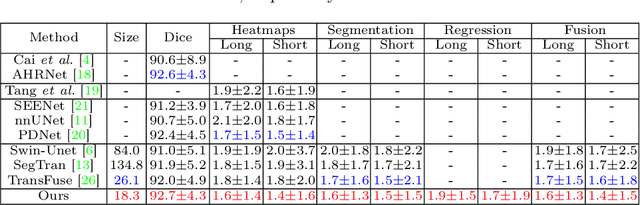
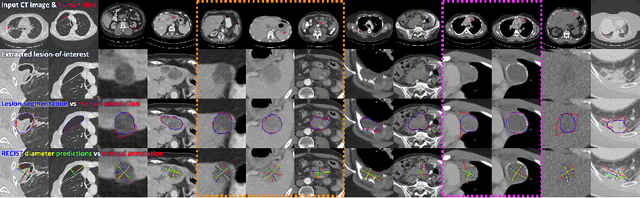
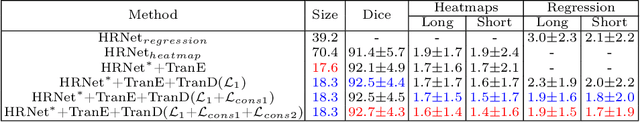
Abstract:Automatically measuring lesion/tumor size with RECIST (Response Evaluation Criteria In Solid Tumors) diameters and segmentation is important for computer-aided diagnosis. Although it has been studied in recent years, there is still space to improve its accuracy and robustness, such as (1) enhancing features by incorporating rich contextual information while keeping a high spatial resolution and (2) involving new tasks and losses for joint optimization. To reach this goal, this paper proposes a transformer-based network (MeaFormer, Measurement transFormer) for lesion RECIST diameter prediction and segmentation (LRDPS). It is formulated as three correlative and complementary tasks: lesion segmentation, heatmap prediction, and keypoint regression. To the best of our knowledge, it is the first time to use keypoint regression for RECIST diameter prediction. MeaFormer can enhance high-resolution features by employing transformers to capture their long-range dependencies. Two consistency losses are introduced to explicitly build relationships among these tasks for better optimization. Experiments show that MeaFormer achieves the state-of-the-art performance of LRDPS on the large-scale DeepLesion dataset and produces promising results of two downstream clinic-relevant tasks, i.e., 3D lesion segmentation and RECIST assessment in longitudinal studies.
PieTrack: An MOT solution based on synthetic data training and self-supervised domain adaptation
Jul 22, 2022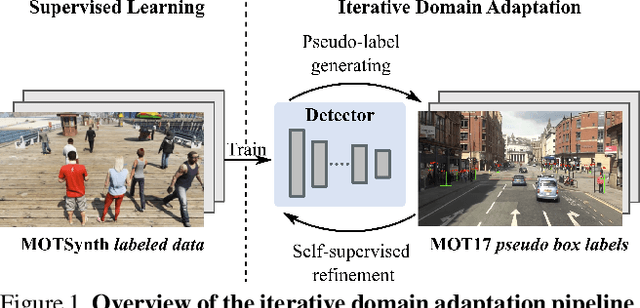
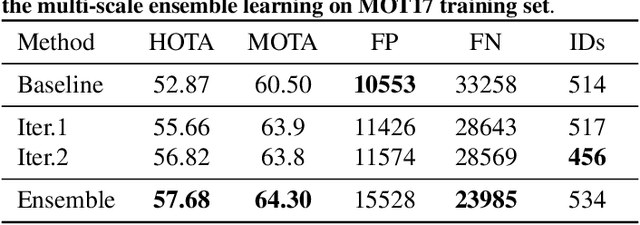
Abstract:In order to cope with the increasing demand for labeling data and privacy issues with human detection, synthetic data has been used as a substitute and showing promising results in human detection and tracking tasks. We participate in the 7th Workshop on Benchmarking Multi-Target Tracking (BMTT), themed on "How Far Can Synthetic Data Take us"? Our solution, PieTrack, is developed based on synthetic data without using any pre-trained weights. We propose a self-supervised domain adaptation method that enables mitigating the domain shift issue between the synthetic (e.g., MOTSynth) and real data (e.g., MOT17) without involving extra human labels. By leveraging the proposed multi-scale ensemble inference, we achieved a final HOTA score of 58.7 on the MOT17 testing set, ranked third place in the challenge.
Deeply-Supervised Density Regression for Automatic Cell Counting in Microscopy Images
Nov 10, 2020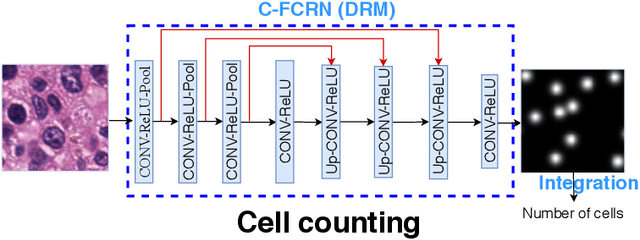
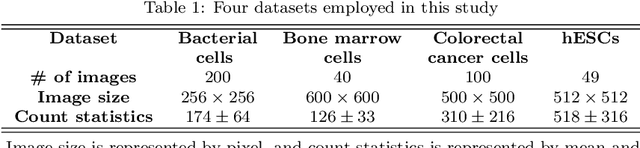
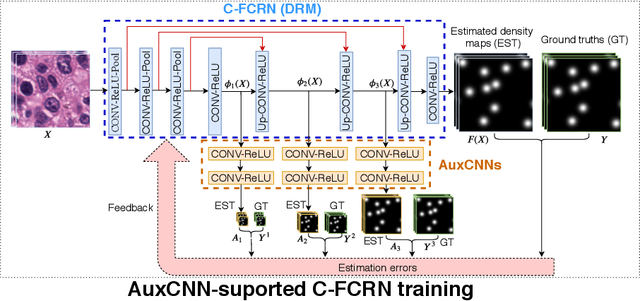
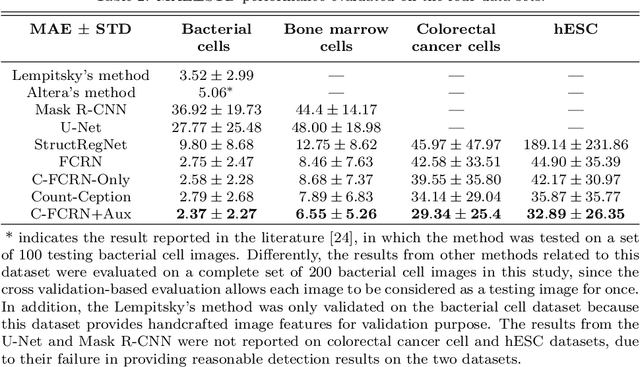
Abstract:Accurately counting the number of cells in microscopy images is required in many medical diagnosis and biological studies. This task is tedious, time-consuming, and prone to subjective errors. However, designing automatic counting methods remains challenging due to low image contrast, complex background, large variance in cell shapes and counts, and significant cell occlusions in two-dimensional microscopy images. In this study, we proposed a new density regression-based method for automatically counting cells in microscopy images. The proposed method processes two innovations compared to other state-of-the-art density regression-based methods. First, the density regression model (DRM) is designed as a concatenated fully convolutional regression network (C-FCRN) to employ multi-scale image features for the estimation of cell density maps from given images. Second, auxiliary convolutional neural networks (AuxCNNs) are employed to assist in the training of intermediate layers of the designed C-FCRN to improve the DRM performance on unseen datasets. Experimental studies evaluated on four datasets demonstrate the superior performance of the proposed method.
Learning Numerical Observers using Unsupervised Domain Adaptation
Feb 22, 2020



Abstract:Medical imaging systems are commonly assessed by use of objective image quality measures. Supervised deep learning methods have been investigated to implement numerical observers for task-based image quality assessment. However, labeling large amounts of experimental data to train deep neural networks is tedious, expensive, and prone to subjective errors. Computer-simulated image data can potentially be employed to circumvent these issues; however, it is often difficult to computationally model complicated anatomical structures, noise sources, and the response of real world imaging systems. Hence, simulated image data will generally possess physical and statistical differences from the experimental image data they seek to emulate. Within the context of machine learning, these differences between the sets of two images is referred to as domain shift. In this study, we propose and investigate the use of an adversarial domain adaptation method to mitigate the deleterious effects of domain shift between simulated and experimental image data for deep learning-based numerical observers (DL-NOs) that are trained on simulated images but applied to experimental ones. In the proposed method, a DL-NO will initially be trained on computer-simulated image data and subsequently adapted for use with experimental image data, without the need for any labeled experimental images. As a proof of concept, a binary signal detection task is considered. The success of this strategy as a function of the degree of domain shift present between the simulated and experimental image data is investigated.
Automatic microscopic cell counting by use of unsupervised adversarial domain adaptation and supervised density regression
Mar 22, 2019Abstract:Accurate cell counting in microscopic images is important for medical diagnoses and biological studies. However, manual cell counting is very time-consuming, tedious, and prone to subjective errors. We propose a new density regression-based method for automatic cell counting that reduces the need to manually annotate experimental images. A supervised learning-based density regression model (DRM) is trained with annotated synthetic images (the source domain) and their corresponding ground truth density maps. A domain adaptation model (DAM) is built to map experimental images (the target domain) to the feature space of the source domain. By use of the unsupervised learning-based DAM and supervised learning-based DRM, a cell density map of a given target image can be estimated, from which the number of cells can be counted. Results from experimental immunofluorescent microscopic images of human embryonic stem cells demonstrate the promising performance of the proposed counting method.
Automatic microscopic cell counting by use of deeply-supervised density regression model
Mar 22, 2019Abstract:Accurately counting cells in microscopic images is important for medical diagnoses and biological studies, but manual cell counting is very tedious, time-consuming, and prone to subjective errors, and automatic counting can be less accurate than desired. To improve the accuracy of automatic cell counting, we propose here a novel method that employs deeply-supervised density regression. A fully convolutional neural network (FCNN) serves as the primary FCNN for density map regression. Innovatively, a set of auxiliary FCNNs are employed to provide additional supervision for learning the intermediate layers of the primary CNN to improve network performance. In addition, the primary CNN is designed as a concatenating framework to integrate multi-scale features through shortcut connections in the network, which improves the granularity of the features extracted from the intermediate CNN layers and further supports the final density map estimation.
Convolutional neural network based automatic plaque characterization from intracoronary optical coherence tomography images
Jul 10, 2018Abstract:Optical coherence tomography (OCT) can provide high-resolution cross-sectional images for analyzing superficial plaques in coronary arteries. Commonly, plaque characterization using intra-coronary OCT images is performed manually by expert observers. This manual analysis is time consuming and its accuracy heavily relies on the experience of human observers. Traditional machine learning based methods, such as the least squares support vector machine and random forest methods, have been recently employed to automatically characterize plaque regions in OCT images. Several processing steps, including feature extraction, informative feature selection, and final pixel classification, are commonly used in these traditional methods. Therefore, the final classification accuracy can be jeopardized by error or inaccuracy within each of these steps. In this study, we proposed a convolutional neural network (CNN) based method to automatically characterize plaques in OCT images. Unlike traditional methods, our method uses the image as a direct input and performs classification as a single-step process. The experiments on 269 OCT images showed that the average prediction accuracy of CNN-based method was 0.866, which indicated a great promise for clinical translation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge