Kyaw Thu Minn
Deeply-Supervised Density Regression for Automatic Cell Counting in Microscopy Images
Nov 10, 2020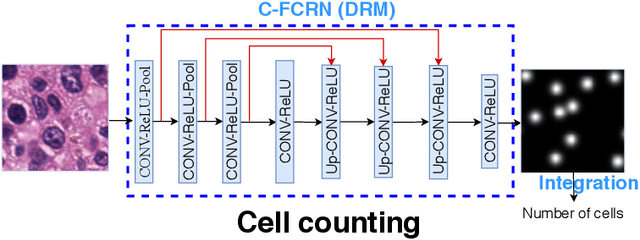
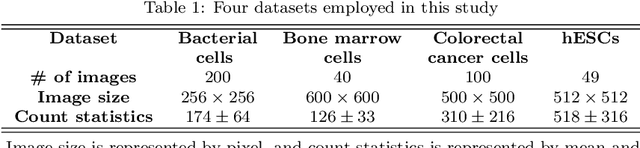
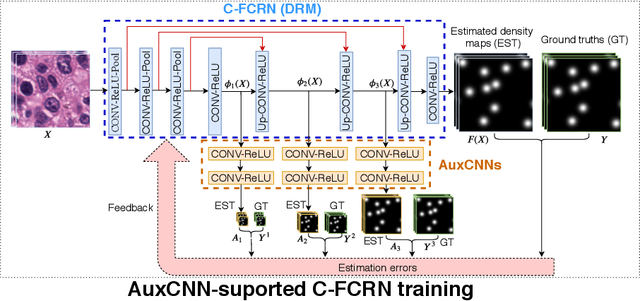
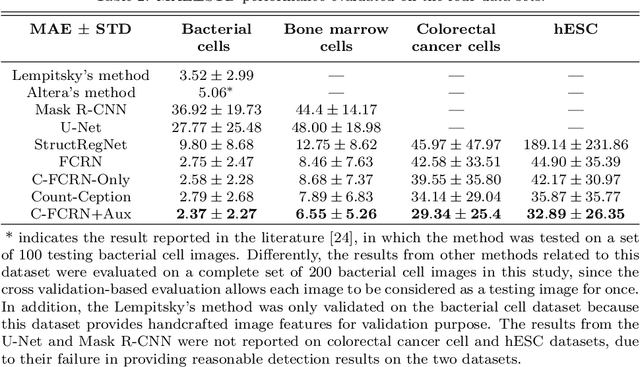
Abstract:Accurately counting the number of cells in microscopy images is required in many medical diagnosis and biological studies. This task is tedious, time-consuming, and prone to subjective errors. However, designing automatic counting methods remains challenging due to low image contrast, complex background, large variance in cell shapes and counts, and significant cell occlusions in two-dimensional microscopy images. In this study, we proposed a new density regression-based method for automatically counting cells in microscopy images. The proposed method processes two innovations compared to other state-of-the-art density regression-based methods. First, the density regression model (DRM) is designed as a concatenated fully convolutional regression network (C-FCRN) to employ multi-scale image features for the estimation of cell density maps from given images. Second, auxiliary convolutional neural networks (AuxCNNs) are employed to assist in the training of intermediate layers of the designed C-FCRN to improve the DRM performance on unseen datasets. Experimental studies evaluated on four datasets demonstrate the superior performance of the proposed method.
Automatic microscopic cell counting by use of unsupervised adversarial domain adaptation and supervised density regression
Mar 22, 2019Abstract:Accurate cell counting in microscopic images is important for medical diagnoses and biological studies. However, manual cell counting is very time-consuming, tedious, and prone to subjective errors. We propose a new density regression-based method for automatic cell counting that reduces the need to manually annotate experimental images. A supervised learning-based density regression model (DRM) is trained with annotated synthetic images (the source domain) and their corresponding ground truth density maps. A domain adaptation model (DAM) is built to map experimental images (the target domain) to the feature space of the source domain. By use of the unsupervised learning-based DAM and supervised learning-based DRM, a cell density map of a given target image can be estimated, from which the number of cells can be counted. Results from experimental immunofluorescent microscopic images of human embryonic stem cells demonstrate the promising performance of the proposed counting method.
Automatic microscopic cell counting by use of deeply-supervised density regression model
Mar 22, 2019Abstract:Accurately counting cells in microscopic images is important for medical diagnoses and biological studies, but manual cell counting is very tedious, time-consuming, and prone to subjective errors, and automatic counting can be less accurate than desired. To improve the accuracy of automatic cell counting, we propose here a novel method that employs deeply-supervised density regression. A fully convolutional neural network (FCNN) serves as the primary FCNN for density map regression. Innovatively, a set of auxiliary FCNNs are employed to provide additional supervision for learning the intermediate layers of the primary CNN to improve network performance. In addition, the primary CNN is designed as a concatenating framework to integrate multi-scale features through shortcut connections in the network, which improves the granularity of the features extracted from the intermediate CNN layers and further supports the final density map estimation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge