Mark A. Anastasio
A Learning-based Framework for Spatial Impulse Response Compensation in 3D Photoacoustic Computed Tomography
Jan 28, 2026Abstract:Photoacoustic computed tomography (PACT) is a promising imaging modality that combines the advantages of optical contrast with ultrasound detection. Utilizing ultrasound transducers with larger surface areas can improve detection sensitivity. However, when computationally efficient analytic reconstruction methods that neglect the spatial impulse responses (SIRs) of the transducer are employed, the spatial resolution of the reconstructed images will be compromised. Although optimization-based reconstruction methods can explicitly account for SIR effects, their computational cost is generally high, particularly in three-dimensional (3D) applications. To address the need for accurate but rapid 3D PACT image reconstruction, this study presents a framework for establishing a learned SIR compensation method that operates in the data domain. The learned compensation method maps SIR-corrupted PACT measurement data to compensated data that would have been recorded by idealized point-like transducers. Subsequently, the compensated data can be used with a computationally efficient reconstruction method that neglects SIR effects. Two variants of the learned compensation model are investigated that employ a U-Net model and a specifically designed, physics-inspired model, referred to as Deconv-Net. A fast and analytical training data generation procedure is also a component of the presented framework. The framework is rigorously validated in virtual imaging studies, demonstrating resolution improvement and robustness to noise variations, object complexity, and sound speed heterogeneity. When applied to in-vivo breast imaging data, the learned compensation models revealed fine structures that had been obscured by SIR-induced artifacts. To our knowledge, this is the first demonstration of learned SIR compensation in 3D PACT imaging.
Benchmarking Deep Learning-Based Reconstruction Methods for Photoacoustic Computed Tomography with Clinically Relevant Synthetic Datasets
Jan 23, 2026Abstract:Deep learning (DL)-based image reconstruction methods for photoacoustic computed tomography (PACT) have developed rapidly in recent years. However, most existing methods have not employed standardized datasets, and their evaluations rely on traditional image quality (IQ) metrics that may lack clinical relevance. The absence of a standardized framework for clinically meaningful IQ assessment hinders fair comparison and raises concerns about the reproducibility and reliability of reported advancements in PACT. A benchmarking framework is proposed that provides open-source, anatomically plausible synthetic datasets and evaluation strategies for DL-based acoustic inversion methods in PACT. The datasets each include over 11,000 two-dimensional (2D) stochastic breast objects with clinically relevant lesions and paired measurements at varying modeling complexity. The evaluation strategies incorporate both traditional and task-based IQ measures to assess fidelity and clinical utility. A preliminary benchmarking study is conducted to demonstrate the framework's utility by comparing DL-based and physics-based reconstruction methods. The benchmarking study demonstrated that the proposed framework enabled comprehensive, quantitative comparisons of reconstruction performance and revealed important limitations in certain DL-based methods. Although they performed well according to traditional IQ measures, they often failed to accurately recover lesions. This highlights the inadequacy of traditional metrics and motivates the need for task-based assessments. The proposed benchmarking framework enables systematic comparisons of DL-based acoustic inversion methods for 2D PACT. By integrating clinically relevant synthetic datasets with rigorous evaluation protocols, it enables reproducible, objective assessments and facilitates method development and system optimization in PACT.
On the Utility of Virtual Staining for Downstream Applications as it relates to Task Network Capacity
Jul 31, 2025Abstract:Virtual staining, or in-silico-labeling, has been proposed to computationally generate synthetic fluorescence images from label-free images by use of deep learning-based image-to-image translation networks. In most reported studies, virtually stained images have been assessed only using traditional image quality measures such as structural similarity or signal-to-noise ratio. However, in biomedical imaging, images are typically acquired to facilitate an image-based inference, which we refer to as a downstream biological or clinical task. This study systematically investigates the utility of virtual staining for facilitating clinically relevant downstream tasks (like segmentation or classification) with consideration of the capacity of the deep neural networks employed to perform the tasks. Comprehensive empirical evaluations were conducted using biological datasets, assessing task performance by use of label-free, virtually stained, and ground truth fluorescence images. The results demonstrated that the utility of virtual staining is largely dependent on the ability of the segmentation or classification task network to extract meaningful task-relevant information, which is related to the concept of network capacity. Examples are provided in which virtual staining does not improve, or even degrades, segmentation or classification performance when the capacity of the associated task network is sufficiently large. The results demonstrate that task network capacity should be considered when deciding whether to perform virtual staining.
Learned Correction Methods for Ultrasound Computed Tomography Imaging Using Simplified Physics Models
Feb 13, 2025


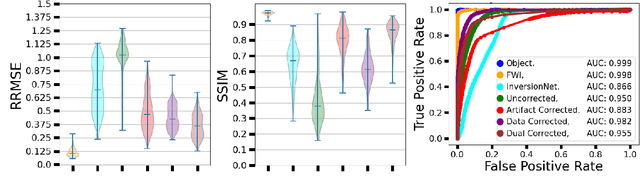
Abstract:Ultrasound computed tomography (USCT) is an emerging modality for breast imaging. Image reconstruction methods that incorporate accurate wave physics produce high resolution quantitative images of acoustic properties but are computationally expensive. The use of a simplified linear model in reconstruction reduces computational expense at the cost of reduced accuracy. This work aims to systematically compare different learning approaches for USCT reconstruction utilizing simplified linear models. This work considered various learning approaches to compensate for errors stemming from a linearized wave propagation model: correction in the data and image domains. The resulting image reconstruction methods are systematically assessed, alongside data-driven and model-based methods, in four virtual imaging studies utilizing anatomically realistic numerical phantoms. Image quality was assessed utilizing relative root mean square error (RRMSE), structural similarity index measure (SSIM), and a task-based assessment for tumor detection. Correction in the measurement domain resulted in images with minor visual artifacts and highly accurate task performance. Correction in the image domain demonstrated a heavy bias on training data, resulting in hallucinations, but greater robustness to measurement noise. Combining both forms of correction performed best in terms of RRMSE and SSIM, at the cost of task performance. This work systematically assessed learned reconstruction methods incorporating an approximated physical model for USCT imaging. Results demonstrated the importance of incorporating physics, compared to data-driven methods. Learning a correction in the data domain led to better task performance and robust out-of-distribution generalization compared to correction in the image domain.
Estimating Task-based Performance Bounds for Accelerated MRI Image Reconstruction Methods by Use of Learned-Ideal Observers
Jan 16, 2025



Abstract:Medical imaging systems are commonly assessed and optimized by the use of objective measures of image quality (IQ). The performance of the ideal observer (IO) acting on imaging measurements has long been advocated as a figure-of-merit to guide the optimization of imaging systems. For computed imaging systems, the performance of the IO acting on imaging measurements also sets an upper bound on task-performance that no image reconstruction method can transcend. As such, estimation of IO performance can provide valuable guidance when designing under-sampled data-acquisition techniques by enabling the identification of designs that will not permit the reconstruction of diagnostically inappropriate images for a specified task - no matter how advanced the reconstruction method is or how plausible the reconstructed images appear. The need for such analysis is urgent because of the substantial increase of medical device submissions on deep learning-based image reconstruction methods and the fact that they may produce clean images disguising the potential loss of diagnostic information when data is aggressively under-sampled. Recently, convolutional neural network (CNN) approximated IOs (CNN-IOs) was investigated for estimating the performance of data space IOs to establish task-based performance bounds for image reconstruction, under an X-ray computed tomographic (CT) context. In this work, the application of such data space CNN-IO analysis to multi-coil magnetic resonance imaging (MRI) systems has been explored. This study utilized stylized multi-coil sensitivity encoding (SENSE) MRI systems and deep-generated stochastic brain models to demonstrate the approach. Signal-known-statistically and background-known-statistically (SKS/BKS) binary signal detection tasks were selected to study the impact of different acceleration factors on the data space IO performance.
Learning a Filtered Backprojection Reconstruction Method for Photoacoustic Computed Tomography with Hemispherical Measurement Geometries
Dec 02, 2024Abstract:In certain three-dimensional (3D) applications of photoacoustic computed tomography (PACT), including \textit{in vivo} breast imaging, hemispherical measurement apertures that enclose the object within their convex hull are employed for data acquisition. Data acquired with such measurement geometries are referred to as \textit{half-scan} data, as only half of a complete spherical measurement aperture is employed. Although previous studies have demonstrated that half-scan data can uniquely and stably reconstruct the sought-after object, no closed-form reconstruction formula for use with half-scan data has been reported. To address this, a semi-analytic reconstruction method in the form of filtered backprojection (FBP), referred to as the half-scan FBP method, is developed in this work. Because the explicit form of the filtering operation in the half-scan FBP method is not currently known, a learning-based method is proposed to approximate it. The proposed method is systematically investigated by use of virtual imaging studies of 3D breast PACT that employ ensembles of numerical breast phantoms and a physics-based model of the data acquisition process. The method is subsequently applied to experimental data acquired in an \textit{in vivo} breast PACT study. The results confirm that the half-scan FBP method can accurately reconstruct 3D images from half-scan data. Importantly, because the sought-after inverse mapping is well-posed, the reconstruction method remains accurate even when applied to data that differ considerably from those employed to learn the filtering operation.
Physics and Deep Learning in Computational Wave Imaging
Oct 10, 2024



Abstract:Computational wave imaging (CWI) extracts hidden structure and physical properties of a volume of material by analyzing wave signals that traverse that volume. Applications include seismic exploration of the Earth's subsurface, acoustic imaging and non-destructive testing in material science, and ultrasound computed tomography in medicine. Current approaches for solving CWI problems can be divided into two categories: those rooted in traditional physics, and those based on deep learning. Physics-based methods stand out for their ability to provide high-resolution and quantitatively accurate estimates of acoustic properties within the medium. However, they can be computationally intensive and are susceptible to ill-posedness and nonconvexity typical of CWI problems. Machine learning-based computational methods have recently emerged, offering a different perspective to address these challenges. Diverse scientific communities have independently pursued the integration of deep learning in CWI. This review delves into how contemporary scientific machine-learning (ML) techniques, and deep neural networks in particular, have been harnessed to tackle CWI problems. We present a structured framework that consolidates existing research spanning multiple domains, including computational imaging, wave physics, and data science. This study concludes with important lessons learned from existing ML-based methods and identifies technical hurdles and emerging trends through a systematic analysis of the extensive literature on this topic.
Revisiting the joint estimation of initial pressure and speed-of-sound distributions in photoacoustic computed tomography with consideration of canonical object constraints
Oct 05, 2024Abstract:In photoacoustic computed tomography (PACT) the accurate estimation of the initial pressure (IP) distribution generally requires knowledge of the object's heterogeneous speed-of-sound (SOS) distribution. Although hybrid imagers that combine ultrasound tomography with PACT have been proposed, in many current applications of PACT the SOS distribution remains unknown. Joint reconstruction (JR) of the IP and SOS distributions from PACT measurement data alone can address this issue. However, this joint estimation problem is ill-posed and corresponds to a non-convex optimization problem. While certain regularization strategies have been deployed, stabilizing the JR problem to yield accurate estimates of the IP and SOS distributions has remained an open challenge. To address this, the presented numerical studies explore the effectiveness of easy to implement canonical object constraints for stabilizing the JR problem. The considered constraints include support, bound, and total variation constraints, which are incorporated into an optimization-based method for JR. Computer-simulation studies that employ anatomically realistic numerical breast phantoms are conducted to evaluate the impact of these object constraints on JR accuracy. Additionally, the impact of certain data inconsistencies, such as caused by measurement noise and physics modeling mismatches, on the effectiveness of the object constraints is investigated. The results demonstrate, for the first time, that the incorporation of canonical object constraints in an optimization-based image reconstruction method holds significant potential for mitigating the ill-posed nature of the PACT JR problem.
Identifying Functional Brain Networks of Spatiotemporal Wide-Field Calcium Imaging Data via a Long Short-Term Memory Autoencoder
May 30, 2024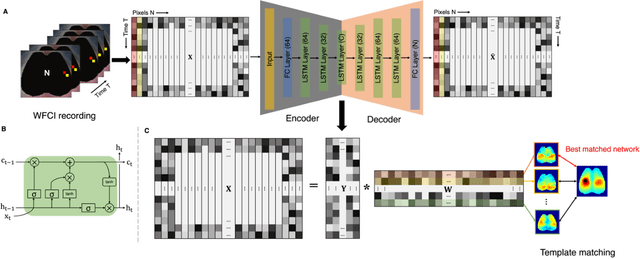
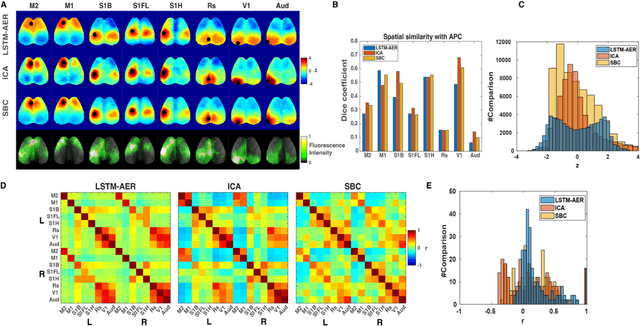
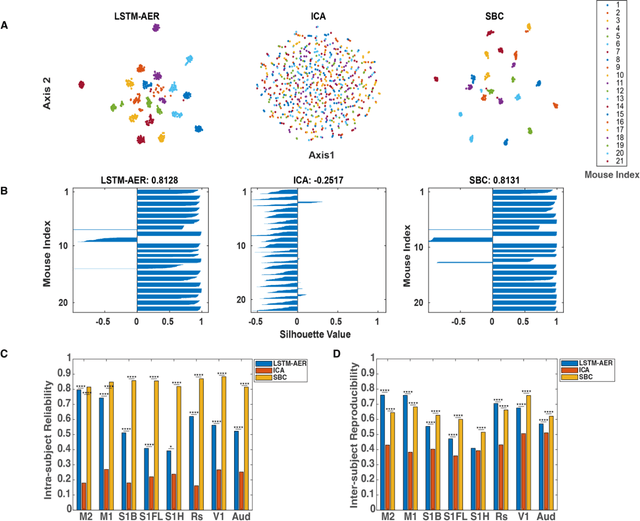
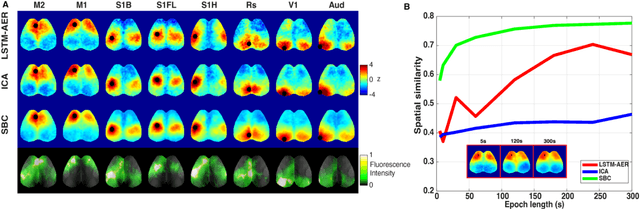
Abstract:Wide-field calcium imaging (WFCI) that records neural calcium dynamics allows for identification of functional brain networks (FBNs) in mice that express genetically encoded calcium indicators. Estimating FBNs from WFCI data is commonly achieved by use of seed-based correlation (SBC) analysis and independent component analysis (ICA). These two methods are conceptually distinct and each possesses limitations. Recent success of unsupervised representation learning in neuroimage analysis motivates the investigation of such methods to identify FBNs. In this work, a novel approach referred as LSTM-AER, is proposed in which a long short-term memory (LSTM) autoencoder (AE) is employed to learn spatial-temporal latent embeddings from WFCI data, followed by an ordinary least square regression (R) to estimate FBNs. The goal of this study is to elucidate and illustrate, qualitatively and quantitatively, the FBNs identified by use of the LSTM-AER method and compare them to those from traditional SBC and ICA. It was observed that spatial FBN maps produced from LSTM-AER resembled those derived by SBC and ICA while better accounting for intra-subject variation, data from a single hemisphere, shorter epoch lengths and tunable number of latent components. The results demonstrate the potential of unsupervised deep learning-based approaches to identifying and mapping FBNs.
Prior-guided Diffusion Model for Cell Segmentation in Quantitative Phase Imaging
May 10, 2024



Abstract:Purpose: Quantitative phase imaging (QPI) is a label-free technique that provides high-contrast images of tissues and cells without the use of chemicals or dyes. Accurate semantic segmentation of cells in QPI is essential for various biomedical applications. While DM-based segmentation has demonstrated promising results, the requirement for multiple sampling steps reduces efficiency. This study aims to enhance DM-based segmentation by introducing prior-guided content information into the starting noise, thereby minimizing inefficiencies associated with multiple sampling. Approach: A prior-guided mechanism is introduced into DM-based segmentation, replacing randomly sampled starting noise with noise informed by content information. This mechanism utilizes another trained DM and DDIM inversion to incorporate content information from the to-be-segmented images into the starting noise. An evaluation method is also proposed to assess the quality of the starting noise, considering both content and distribution information. Results: Extensive experiments on various QPI datasets for cell segmentation showed that the proposed method achieved superior performance in DM-based segmentation with only a single sampling. Ablation studies and visual analysis further highlighted the significance of content priors in DM-based segmentation. Conclusion: The proposed method effectively leverages prior content information to improve DM-based segmentation, providing accurate results while reducing the need for multiple samplings. The findings emphasize the importance of integrating content priors into DM-based segmentation methods for optimal performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge