Rucha Deshpande
Knowledge-based anomaly detection for identifying network-induced shape artifacts
Nov 06, 2025Abstract:Synthetic data provides a promising approach to address data scarcity for training machine learning models; however, adoption without proper quality assessments may introduce artifacts, distortions, and unrealistic features that compromise model performance and clinical utility. This work introduces a novel knowledge-based anomaly detection method for detecting network-induced shape artifacts in synthetic images. The introduced method utilizes a two-stage framework comprising (i) a novel feature extractor that constructs a specialized feature space by analyzing the per-image distribution of angle gradients along anatomical boundaries, and (ii) an isolation forest-based anomaly detector. We demonstrate the effectiveness of the method for identifying network-induced shape artifacts in two synthetic mammography datasets from models trained on CSAW-M and VinDr-Mammo patient datasets respectively. Quantitative evaluation shows that the method successfully concentrates artifacts in the most anomalous partition (1st percentile), with AUC values of 0.97 (CSAW-syn) and 0.91 (VMLO-syn). In addition, a reader study involving three imaging scientists confirmed that images identified by the method as containing network-induced shape artifacts were also flagged by human readers with mean agreement rates of 66% (CSAW-syn) and 68% (VMLO-syn) for the most anomalous partition, approximately 1.5-2 times higher than the least anomalous partition. Kendall-Tau correlations between algorithmic and human rankings were 0.45 and 0.43 for the two datasets, indicating reasonable agreement despite the challenging nature of subtle artifact detection. This method is a step forward in the responsible use of synthetic data, as it allows developers to evaluate synthetic images for known anatomic constraints and pinpoint and address specific issues to improve the overall quality of a synthetic dataset.
Evaluation of Machine-generated Biomedical Images via A Tally-based Similarity Measure
Mar 28, 2025



Abstract:Super-resolution, in-painting, whole-image generation, unpaired style-transfer, and network-constrained image reconstruction each include an aspect of machine-learned image synthesis where the actual ground truth is not known at time of use. It is generally difficult to quantitatively and authoritatively evaluate the quality of synthetic images; however, in mission-critical biomedical scenarios robust evaluation is paramount. In this work, all practical image-to-image comparisons really are relative qualifications, not absolute difference quantifications; and, therefore, meaningful evaluation of generated image quality can be accomplished using the Tversky Index, which is a well-established measure for assessing perceptual similarity. This evaluation procedure is developed and then demonstrated using multiple image data sets, both real and simulated. The main result is that when the subjectivity and intrinsic deficiencies of any feature-encoding choice are put upfront, Tversky's method leads to intuitive results, whereas traditional methods based on summarizing distances in deep feature spaces do not.
Report on the AAPM Grand Challenge on deep generative modeling for learning medical image statistics
May 03, 2024



Abstract:The findings of the 2023 AAPM Grand Challenge on Deep Generative Modeling for Learning Medical Image Statistics are reported in this Special Report. The goal of this challenge was to promote the development of deep generative models (DGMs) for medical imaging and to emphasize the need for their domain-relevant assessment via the analysis of relevant image statistics. As part of this Grand Challenge, a training dataset was developed based on 3D anthropomorphic breast phantoms from the VICTRE virtual imaging toolbox. A two-stage evaluation procedure consisting of a preliminary check for memorization and image quality (based on the Frechet Inception distance (FID)), and a second stage evaluating the reproducibility of image statistics corresponding to domain-relevant radiomic features was developed. A summary measure was employed to rank the submissions. Additional analyses of submissions was performed to assess DGM performance specific to individual feature families, and to identify various artifacts. 58 submissions from 12 unique users were received for this Challenge. The top-ranked submission employed a conditional latent diffusion model, whereas the joint runners-up employed a generative adversarial network, followed by another network for image superresolution. We observed that the overall ranking of the top 9 submissions according to our evaluation method (i) did not match the FID-based ranking, and (ii) differed with respect to individual feature families. Another important finding from our additional analyses was that different DGMs demonstrated similar kinds of artifacts. This Grand Challenge highlighted the need for domain-specific evaluation to further DGM design as well as deployment. It also demonstrated that the specification of a DGM may differ depending on its intended use.
Assessing the capacity of a denoising diffusion probabilistic model to reproduce spatial context
Sep 19, 2023



Abstract:Diffusion models have emerged as a popular family of deep generative models (DGMs). In the literature, it has been claimed that one class of diffusion models -- denoising diffusion probabilistic models (DDPMs) -- demonstrate superior image synthesis performance as compared to generative adversarial networks (GANs). To date, these claims have been evaluated using either ensemble-based methods designed for natural images, or conventional measures of image quality such as structural similarity. However, there remains an important need to understand the extent to which DDPMs can reliably learn medical imaging domain-relevant information, which is referred to as `spatial context' in this work. To address this, a systematic assessment of the ability of DDPMs to learn spatial context relevant to medical imaging applications is reported for the first time. A key aspect of the studies is the use of stochastic context models (SCMs) to produce training data. In this way, the ability of the DDPMs to reliably reproduce spatial context can be quantitatively assessed by use of post-hoc image analyses. Error-rates in DDPM-generated ensembles are reported, and compared to those corresponding to a modern GAN. The studies reveal new and important insights regarding the capacity of DDPMs to learn spatial context. Notably, the results demonstrate that DDPMs hold significant capacity for generating contextually correct images that are `interpolated' between training samples, which may benefit data-augmentation tasks in ways that GANs cannot.
AmbientFlow: Invertible generative models from incomplete, noisy measurements
Sep 09, 2023Abstract:Generative models have gained popularity for their potential applications in imaging science, such as image reconstruction, posterior sampling and data sharing. Flow-based generative models are particularly attractive due to their ability to tractably provide exact density estimates along with fast, inexpensive and diverse samples. Training such models, however, requires a large, high quality dataset of objects. In applications such as computed imaging, it is often difficult to acquire such data due to requirements such as long acquisition time or high radiation dose, while acquiring noisy or partially observed measurements of these objects is more feasible. In this work, we propose AmbientFlow, a framework for learning flow-based generative models directly from noisy and incomplete data. Using variational Bayesian methods, a novel framework for establishing flow-based generative models from noisy, incomplete data is proposed. Extensive numerical studies demonstrate the effectiveness of AmbientFlow in correctly learning the object distribution. The utility of AmbientFlow in a downstream inference task of image reconstruction is demonstrated.
Investigating the robustness of a learning-based method for quantitative phase retrieval from propagation-based x-ray phase contrast measurements under laboratory conditions
Nov 02, 2022Abstract:Quantitative phase retrieval (QPR) in propagation-based x-ray phase contrast imaging of heterogeneous and structurally complicated objects is challenging under laboratory conditions due to partial spatial coherence and polychromaticity. A learning-based method (LBM) provides a non-linear approach to this problem while not being constrained by restrictive assumptions about object properties and beam coherence. In this work, a LBM was assessed for its applicability under practical scenarios by evaluating its robustness and generalizability under typical experimental variations. Towards this end, an end-to-end LBM was employed for QPR under laboratory conditions and its robustness was investigated across various system and object conditions. The robustness of the method was tested via varying propagation distances and its generalizability with respect to object structure and experimental data was also tested. Although the LBM was stable under the studied variations, its successful deployment was found to be affected by choices pertaining to data pre-processing, network training considerations and system modeling. To our knowledge, we demonstrated for the first time, the potential applicability of an end-to-end learning-based quantitative phase retrieval method, trained on simulated data, to experimental propagation-based x-ray phase contrast measurements acquired under laboratory conditions. We considered conditions of polychromaticity, partial spatial coherence, and high noise levels, typical to laboratory conditions. This work further explored the robustness of this method to practical variations in propagation distances and object structure with the goal of assessing its potential for experimental use. Such an exploration of any LBM (irrespective of its network architecture) before practical deployment provides an understanding of its potential behavior under experimental settings.
A Method for Evaluating the Capacity of Generative Adversarial Networks to Reproduce High-order Spatial Context
Nov 24, 2021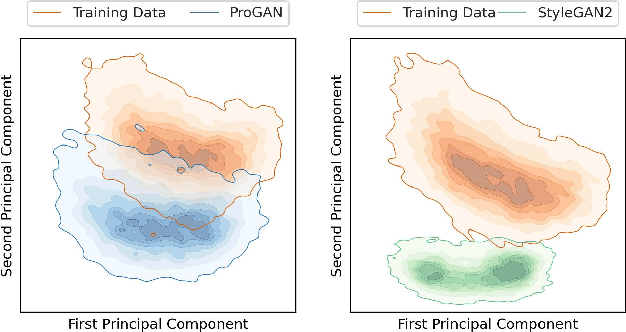
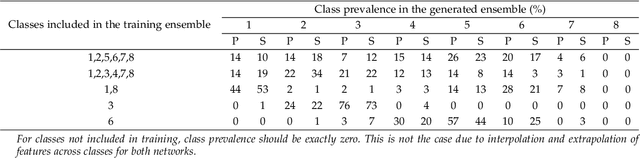
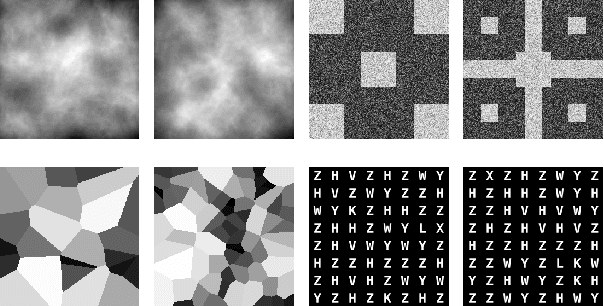
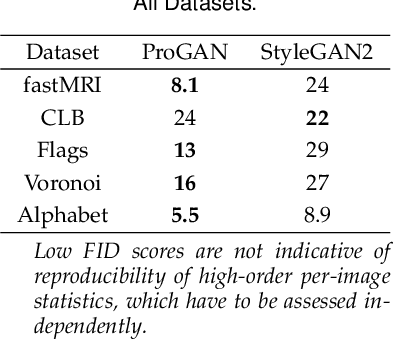
Abstract:Generative adversarial networks are a kind of deep generative model with the potential to revolutionize biomedical imaging. This is because GANs have a learned capacity to draw whole-image variates from a lower-dimensional representation of an unknown, high-dimensional distribution that fully describes the input training images. The overarching problem with GANs in clinical applications is that there is not adequate or automatic means of assessing the diagnostic quality of images generated by GANs. In this work, we demonstrate several tests of the statistical accuracy of images output by two popular GAN architectures. We designed several stochastic object models (SOMs) of distinct features that can be recovered after generation by a trained GAN. Several of these features are high-order, algorithmic pixel-arrangement rules which are not readily expressed in covariance matrices. We designed and validated statistical classifiers to detect the known arrangement rules. We then tested the rates at which the different GANs correctly reproduced the rules under a variety of training scenarios and degrees of feature-class similarity. We found that ensembles of generated images can appear accurate visually, and correspond to low Frechet Inception Distance scores (FID), while not exhibiting the known spatial arrangements. Furthermore, GANs trained on a spectrum of distinct spatial orders did not respect the given prevalence of those orders in the training data. The main conclusion is that while low-order ensemble statistics are largely correct, there are numerous quantifiable errors per image that plausibly can affect subsequent use of the GAN-generated images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge