Prabhat Kc
Estimating Task-based Performance Bounds for Accelerated MRI Image Reconstruction Methods by Use of Learned-Ideal Observers
Jan 16, 2025



Abstract:Medical imaging systems are commonly assessed and optimized by the use of objective measures of image quality (IQ). The performance of the ideal observer (IO) acting on imaging measurements has long been advocated as a figure-of-merit to guide the optimization of imaging systems. For computed imaging systems, the performance of the IO acting on imaging measurements also sets an upper bound on task-performance that no image reconstruction method can transcend. As such, estimation of IO performance can provide valuable guidance when designing under-sampled data-acquisition techniques by enabling the identification of designs that will not permit the reconstruction of diagnostically inappropriate images for a specified task - no matter how advanced the reconstruction method is or how plausible the reconstructed images appear. The need for such analysis is urgent because of the substantial increase of medical device submissions on deep learning-based image reconstruction methods and the fact that they may produce clean images disguising the potential loss of diagnostic information when data is aggressively under-sampled. Recently, convolutional neural network (CNN) approximated IOs (CNN-IOs) was investigated for estimating the performance of data space IOs to establish task-based performance bounds for image reconstruction, under an X-ray computed tomographic (CT) context. In this work, the application of such data space CNN-IO analysis to multi-coil magnetic resonance imaging (MRI) systems has been explored. This study utilized stylized multi-coil sensitivity encoding (SENSE) MRI systems and deep-generated stochastic brain models to demonstrate the approach. Signal-known-statistically and background-known-statistically (SKS/BKS) binary signal detection tasks were selected to study the impact of different acceleration factors on the data space IO performance.
Assessing the performance of CT image denoisers using Laguerre-Gauss Channelized Hotelling Observer for lesion detection
Dec 04, 2024
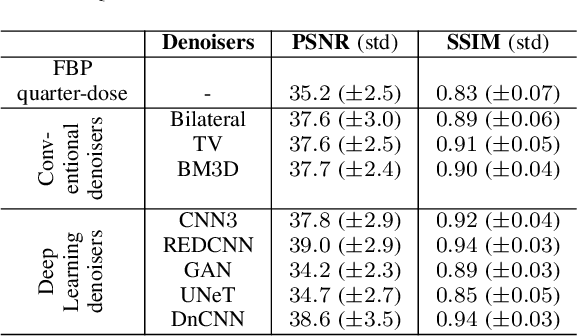
Abstract:The remarkable success of deep learning methods in solving computer vision problems, such as image classification, object detection, scene understanding, image segmentation, etc., has paved the way for their application in biomedical imaging. One such application is in the field of CT image denoising, whereby deep learning methods are proposed to recover denoised images from noisy images acquired at low radiation. Outputs derived from applying deep learning denoising algorithms may appear clean and visually pleasing; however, the underlying diagnostic image quality may not be on par with their normal-dose CT counterparts. In this work, we assessed the image quality of deep learning denoising algorithms by making use of visual perception- and data fidelity-based task-agnostic metrics (like the PSNR and the SSIM) - commonly used in the computer vision - and a task-based detectability assessment (the LCD) - extensively used in the CT imaging. When compared against normal-dose CT images, the deep learning denoisers outperformed low-dose CT based on metrics like the PSNR (by 2.4 to 3.8 dB) and SSIM (by 0.05 to 0.11). However, based on the LCD performance, the detectability using quarter-dose denoised outputs was inferior to that obtained using normal-dose CT scans.
* 2 pages, 2024 IEEE Nuclear Science Symposium (NSS), Medical Imaging Conference (MIC) and Room Temperature Semiconductor Detector Conference (RTSD)
Report on the AAPM Grand Challenge on deep generative modeling for learning medical image statistics
May 03, 2024



Abstract:The findings of the 2023 AAPM Grand Challenge on Deep Generative Modeling for Learning Medical Image Statistics are reported in this Special Report. The goal of this challenge was to promote the development of deep generative models (DGMs) for medical imaging and to emphasize the need for their domain-relevant assessment via the analysis of relevant image statistics. As part of this Grand Challenge, a training dataset was developed based on 3D anthropomorphic breast phantoms from the VICTRE virtual imaging toolbox. A two-stage evaluation procedure consisting of a preliminary check for memorization and image quality (based on the Frechet Inception distance (FID)), and a second stage evaluating the reproducibility of image statistics corresponding to domain-relevant radiomic features was developed. A summary measure was employed to rank the submissions. Additional analyses of submissions was performed to assess DGM performance specific to individual feature families, and to identify various artifacts. 58 submissions from 12 unique users were received for this Challenge. The top-ranked submission employed a conditional latent diffusion model, whereas the joint runners-up employed a generative adversarial network, followed by another network for image superresolution. We observed that the overall ranking of the top 9 submissions according to our evaluation method (i) did not match the FID-based ranking, and (ii) differed with respect to individual feature families. Another important finding from our additional analyses was that different DGMs demonstrated similar kinds of artifacts. This Grand Challenge highlighted the need for domain-specific evaluation to further DGM design as well as deployment. It also demonstrated that the specification of a DGM may differ depending on its intended use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge