A Method for Evaluating the Capacity of Generative Adversarial Networks to Reproduce High-order Spatial Context
Paper and Code
Nov 24, 2021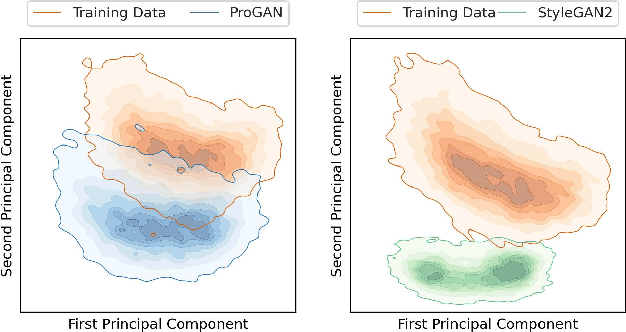
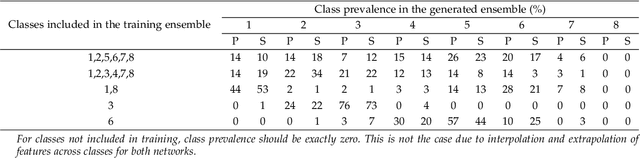
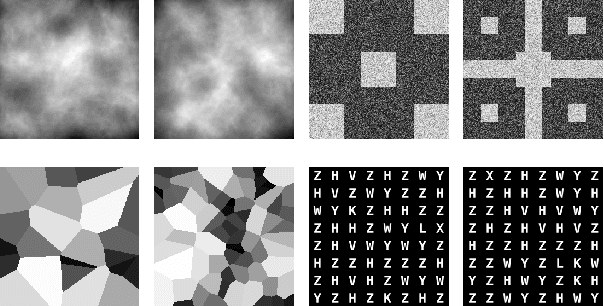
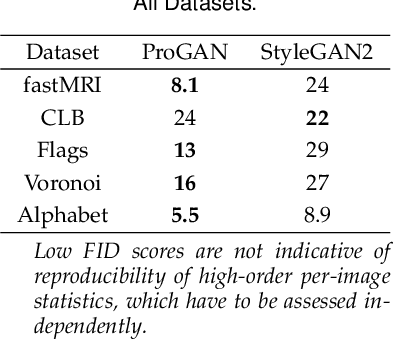
Generative adversarial networks are a kind of deep generative model with the potential to revolutionize biomedical imaging. This is because GANs have a learned capacity to draw whole-image variates from a lower-dimensional representation of an unknown, high-dimensional distribution that fully describes the input training images. The overarching problem with GANs in clinical applications is that there is not adequate or automatic means of assessing the diagnostic quality of images generated by GANs. In this work, we demonstrate several tests of the statistical accuracy of images output by two popular GAN architectures. We designed several stochastic object models (SOMs) of distinct features that can be recovered after generation by a trained GAN. Several of these features are high-order, algorithmic pixel-arrangement rules which are not readily expressed in covariance matrices. We designed and validated statistical classifiers to detect the known arrangement rules. We then tested the rates at which the different GANs correctly reproduced the rules under a variety of training scenarios and degrees of feature-class similarity. We found that ensembles of generated images can appear accurate visually, and correspond to low Frechet Inception Distance scores (FID), while not exhibiting the known spatial arrangements. Furthermore, GANs trained on a spectrum of distinct spatial orders did not respect the given prevalence of those orders in the training data. The main conclusion is that while low-order ensemble statistics are largely correct, there are numerous quantifiable errors per image that plausibly can affect subsequent use of the GAN-generated images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge