Joseph P. Culver
Identifying Functional Brain Networks of Spatiotemporal Wide-Field Calcium Imaging Data via a Long Short-Term Memory Autoencoder
May 30, 2024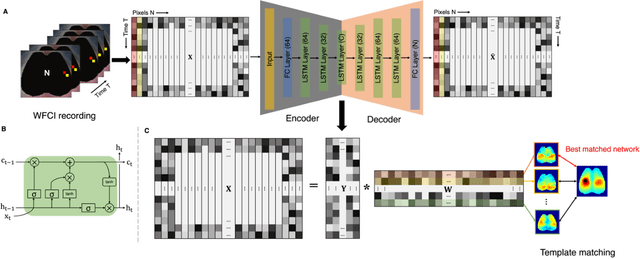
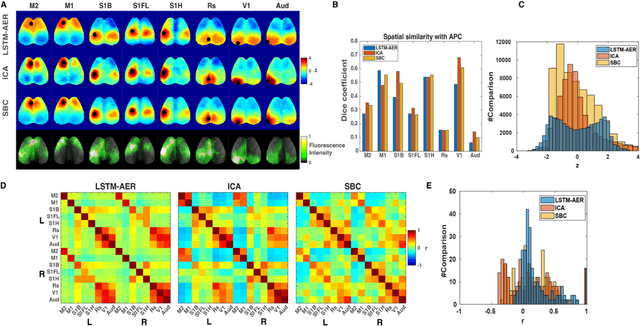
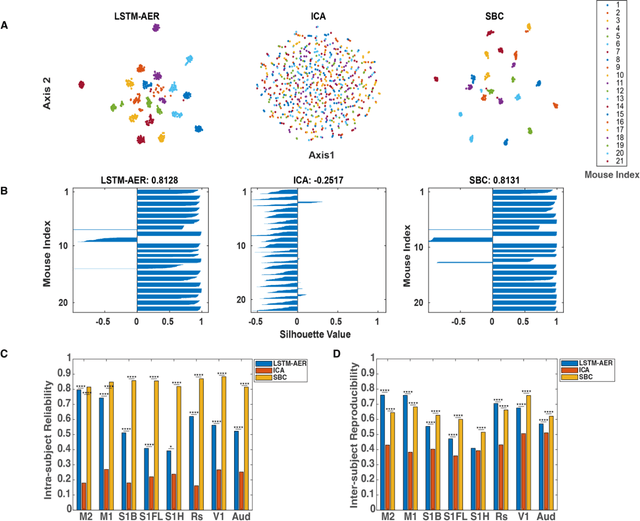
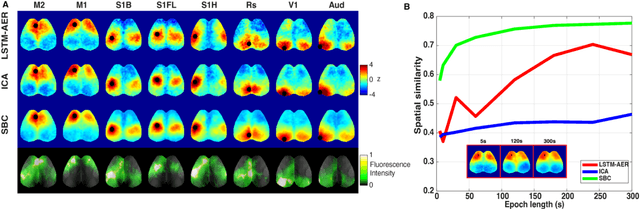
Abstract:Wide-field calcium imaging (WFCI) that records neural calcium dynamics allows for identification of functional brain networks (FBNs) in mice that express genetically encoded calcium indicators. Estimating FBNs from WFCI data is commonly achieved by use of seed-based correlation (SBC) analysis and independent component analysis (ICA). These two methods are conceptually distinct and each possesses limitations. Recent success of unsupervised representation learning in neuroimage analysis motivates the investigation of such methods to identify FBNs. In this work, a novel approach referred as LSTM-AER, is proposed in which a long short-term memory (LSTM) autoencoder (AE) is employed to learn spatial-temporal latent embeddings from WFCI data, followed by an ordinary least square regression (R) to estimate FBNs. The goal of this study is to elucidate and illustrate, qualitatively and quantitatively, the FBNs identified by use of the LSTM-AER method and compare them to those from traditional SBC and ICA. It was observed that spatial FBN maps produced from LSTM-AER resembled those derived by SBC and ICA while better accounting for intra-subject variation, data from a single hemisphere, shorter epoch lengths and tunable number of latent components. The results demonstrate the potential of unsupervised deep learning-based approaches to identifying and mapping FBNs.
Attention-Based CNN-BiLSTM for Sleep State Classification of Spatiotemporal Wide-Field Calcium Imaging Data
Jan 16, 2024Abstract:Background: Wide-field calcium imaging (WFCI) with genetically encoded calcium indicators allows for spatiotemporal recordings of neuronal activity in mice. When applied to the study of sleep, WFCI data are manually scored into the sleep states of wakefulness, non-REM (NREM) and REM by use of adjunct EEG and EMG recordings. However, this process is time-consuming, invasive and often suffers from low inter- and intra-rater reliability. Therefore, an automated sleep state classification method that operates on spatiotemporal WFCI data is desired. New Method: A hybrid network architecture consisting of a convolutional neural network (CNN) to extract spatial features of image frames and a bidirectional long short-term memory network (BiLSTM) with attention mechanism to identify temporal dependencies among different time points was proposed to classify WFCI data into states of wakefulness, NREM and REM sleep. Results: Sleep states were classified with an accuracy of 84% and Cohen's kappa of 0.64. Gradient-weighted class activation maps revealed that the frontal region of the cortex carries more importance when classifying WFCI data into NREM sleep while posterior area contributes most to the identification of wakefulness. The attention scores indicated that the proposed network focuses on short- and long-range temporal dependency in a state-specific manner. Comparison with Existing Method: On a 3-hour WFCI recording, the CNN-BiLSTM achieved a kappa of 0.67, comparable to a kappa of 0.65 corresponding to the human EEG/EMG-based scoring. Conclusions: The CNN-BiLSTM effectively classifies sleep states from spatiotemporal WFCI data and will enable broader application of WFCI in sleep.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge