Zhuchen Shao
Prior-guided Diffusion Model for Cell Segmentation in Quantitative Phase Imaging
May 10, 2024



Abstract:Purpose: Quantitative phase imaging (QPI) is a label-free technique that provides high-contrast images of tissues and cells without the use of chemicals or dyes. Accurate semantic segmentation of cells in QPI is essential for various biomedical applications. While DM-based segmentation has demonstrated promising results, the requirement for multiple sampling steps reduces efficiency. This study aims to enhance DM-based segmentation by introducing prior-guided content information into the starting noise, thereby minimizing inefficiencies associated with multiple sampling. Approach: A prior-guided mechanism is introduced into DM-based segmentation, replacing randomly sampled starting noise with noise informed by content information. This mechanism utilizes another trained DM and DDIM inversion to incorporate content information from the to-be-segmented images into the starting noise. An evaluation method is also proposed to assess the quality of the starting noise, considering both content and distribution information. Results: Extensive experiments on various QPI datasets for cell segmentation showed that the proposed method achieved superior performance in DM-based segmentation with only a single sampling. Ablation studies and visual analysis further highlighted the significance of content priors in DM-based segmentation. Conclusion: The proposed method effectively leverages prior content information to improve DM-based segmentation, providing accurate results while reducing the need for multiple samplings. The findings emphasize the importance of integrating content priors into DM-based segmentation methods for optimal performance.
Semi-Supervised Semantic Segmentation of Cell Nuclei via Diffusion-based Large-Scale Pre-Training and Collaborative Learning
Aug 08, 2023



Abstract:Automated semantic segmentation of cell nuclei in microscopic images is crucial for disease diagnosis and tissue microenvironment analysis. Nonetheless, this task presents challenges due to the complexity and heterogeneity of cells. While supervised deep learning methods are promising, they necessitate large annotated datasets that are time-consuming and error-prone to acquire. Semi-supervised approaches could provide feasible alternatives to this issue. However, the limited annotated data may lead to subpar performance of semi-supervised methods, regardless of the abundance of unlabeled data. In this paper, we introduce a novel unsupervised pre-training-based semi-supervised framework for cell-nuclei segmentation. Our framework is comprised of three main components. Firstly, we pretrain a diffusion model on a large-scale unlabeled dataset. The diffusion model's explicit modeling capability facilitates the learning of semantic feature representation from the unlabeled data. Secondly, we achieve semantic feature aggregation using a transformer-based decoder, where the pretrained diffusion model acts as the feature extractor, enabling us to fully utilize the small amount of labeled data. Finally, we implement a collaborative learning framework between the diffusion-based segmentation model and a supervised segmentation model to further enhance segmentation performance. Experiments were conducted on four publicly available datasets to demonstrate significant improvements compared to competitive semi-supervised segmentation methods and supervised baselines. A series of out-of-distribution tests further confirmed the generality of our framework. Furthermore, thorough ablation experiments and visual analysis confirmed the superiority of our proposed method.
HVTSurv: Hierarchical Vision Transformer for Patient-Level Survival Prediction from Whole Slide Image
Jun 30, 2023
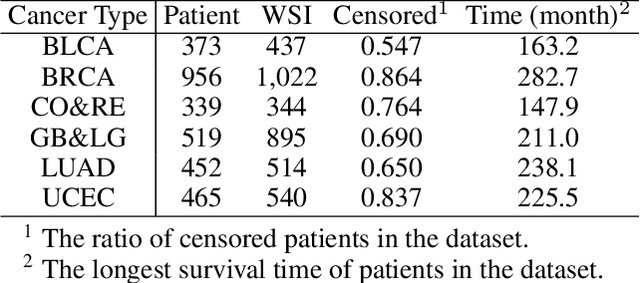
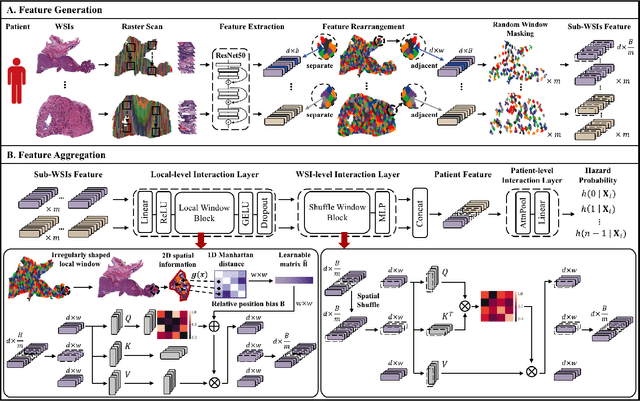
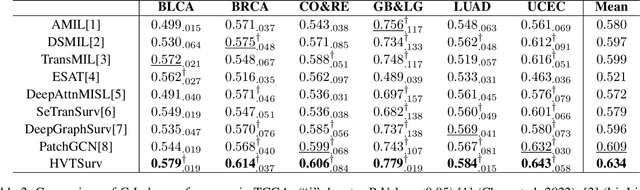
Abstract:Survival prediction based on whole slide images (WSIs) is a challenging task for patient-level multiple instance learning (MIL). Due to the vast amount of data for a patient (one or multiple gigapixels WSIs) and the irregularly shaped property of WSI, it is difficult to fully explore spatial, contextual, and hierarchical interaction in the patient-level bag. Many studies adopt random sampling pre-processing strategy and WSI-level aggregation models, which inevitably lose critical prognostic information in the patient-level bag. In this work, we propose a hierarchical vision Transformer framework named HVTSurv, which can encode the local-level relative spatial information, strengthen WSI-level context-aware communication, and establish patient-level hierarchical interaction. Firstly, we design a feature pre-processing strategy, including feature rearrangement and random window masking. Then, we devise three layers to progressively obtain patient-level representation, including a local-level interaction layer adopting Manhattan distance, a WSI-level interaction layer employing spatial shuffle, and a patient-level interaction layer using attention pooling. Moreover, the design of hierarchical network helps the model become more computationally efficient. Finally, we validate HVTSurv with 3,104 patients and 3,752 WSIs across 6 cancer types from The Cancer Genome Atlas (TCGA). The average C-Index is 2.50-11.30% higher than all the prior weakly supervised methods over 6 TCGA datasets. Ablation study and attention visualization further verify the superiority of the proposed HVTSurv. Implementation is available at: https://github.com/szc19990412/HVTSurv.
AugDiff: Diffusion based Feature Augmentation for Multiple Instance Learning in Whole Slide Image
Mar 11, 2023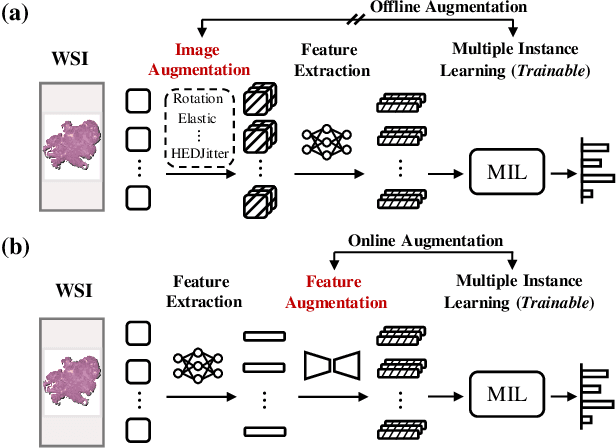
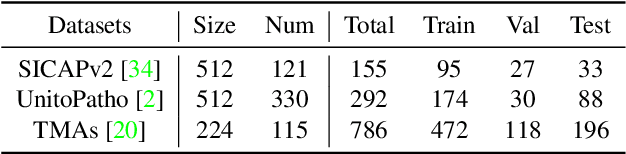
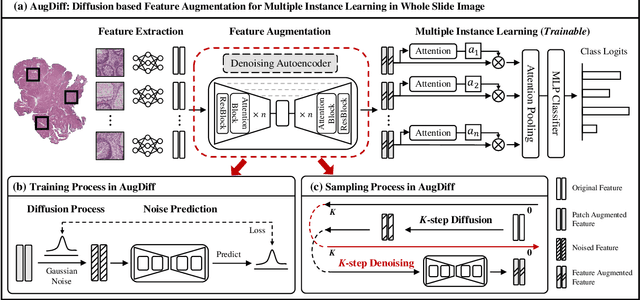
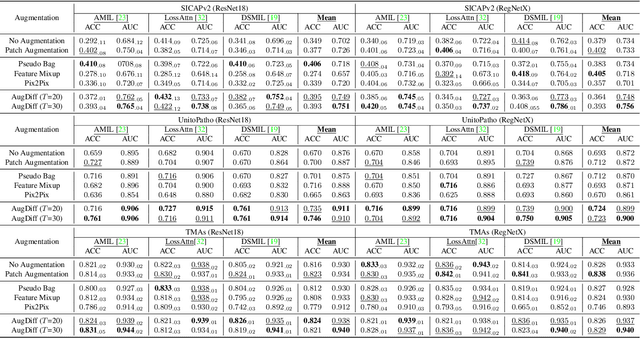
Abstract:Multiple Instance Learning (MIL), a powerful strategy for weakly supervised learning, is able to perform various prediction tasks on gigapixel Whole Slide Images (WSIs). However, the tens of thousands of patches in WSIs usually incur a vast computational burden for image augmentation, limiting the MIL model's improvement in performance. Currently, the feature augmentation-based MIL framework is a promising solution, while existing methods such as Mixup often produce unrealistic features. To explore a more efficient and practical augmentation method, we introduce the Diffusion Model (DM) into MIL for the first time and propose a feature augmentation framework called AugDiff. Specifically, we employ the generation diversity of DM to improve the quality of feature augmentation and the step-by-step generation property to control the retention of semantic information. We conduct extensive experiments over three distinct cancer datasets, two different feature extractors, and three prevalent MIL algorithms to evaluate the performance of AugDiff. Ablation study and visualization further verify the effectiveness. Moreover, we highlight AugDiff's higher-quality augmented feature over image augmentation and its superiority over self-supervised learning. The generalization over external datasets indicates its broader applications.
Multiple Instance Learning with Mixed Supervision in Gleason Grading
Jun 26, 2022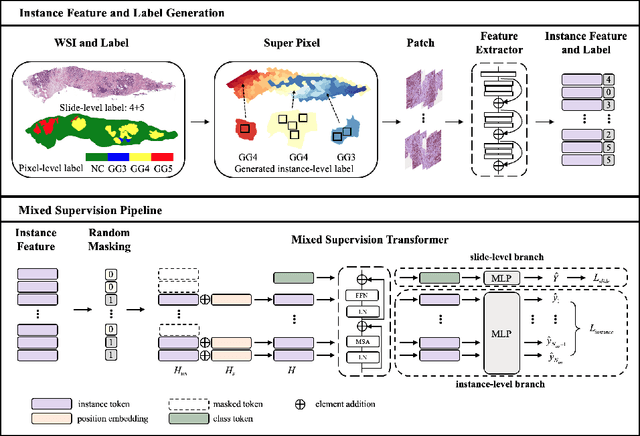
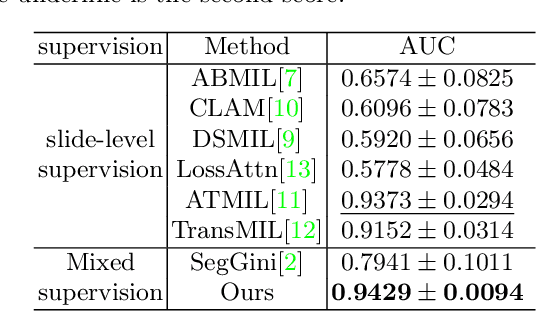
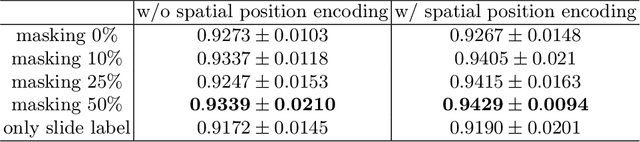
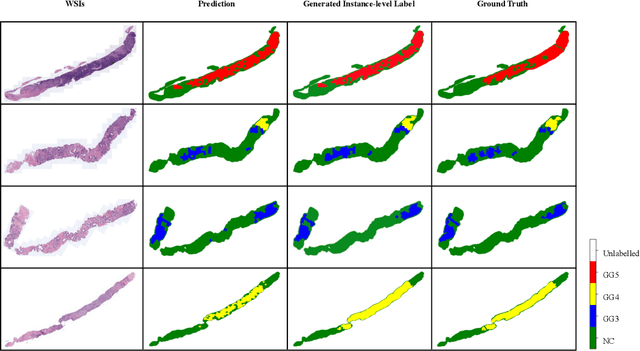
Abstract:With the development of computational pathology, deep learning methods for Gleason grading through whole slide images (WSIs) have excellent prospects. Since the size of WSIs is extremely large, the image label usually contains only slide-level label or limited pixel-level labels. The current mainstream approach adopts multi-instance learning to predict Gleason grades. However, some methods only considering the slide-level label ignore the limited pixel-level labels containing rich local information. Furthermore, the method of additionally considering the pixel-level labels ignores the inaccuracy of pixel-level labels. To address these problems, we propose a mixed supervision Transformer based on the multiple instance learning framework. The model utilizes both slide-level label and instance-level labels to achieve more accurate Gleason grading at the slide level. The impact of inaccurate instance-level labels is further reduced by introducing an efficient random masking strategy in the mixed supervision training process. We achieve the state-of-the-art performance on the SICAPv2 dataset, and the visual analysis shows the accurate prediction results of instance level. The source code is available at https://github.com/bianhao123/Mixed_supervision.
TransMIL: Transformer based Correlated Multiple Instance Learning for Whole Slide Image Classication
Jun 02, 2021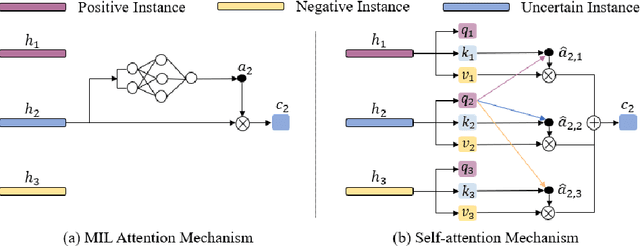
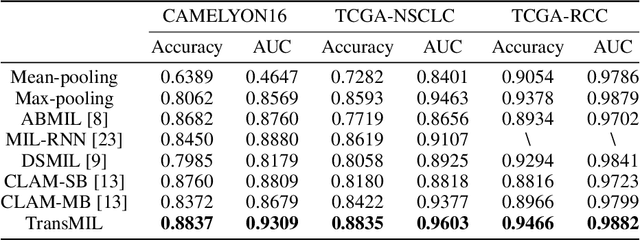
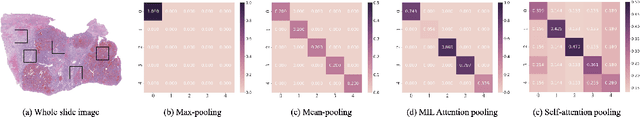
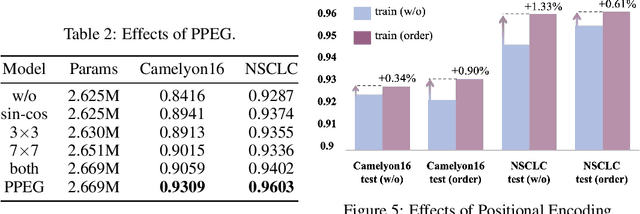
Abstract:Multiple instance learning (MIL) is a powerful tool to solve the weakly supervised classification in whole slide image (WSI) based pathology diagnosis. However, the current MIL methods are usually based on independent and identical distribution hypothesis, thus neglect the correlation among different instances. To address this problem, we proposed a new framework, called correlated MIL, and provided a proof for convergence. Based on this framework, we devised a Transformer based MIL (TransMIL), which explored both morphological and spatial information. The proposed TransMIL can effectively deal with unbalanced/balanced and binary/multiple classification with great visualization and interpretability. We conducted various experiments for three different computational pathology problems and achieved better performance and faster convergence compared with state-of-the-art methods. The test AUC for the binary tumor classification can be up to 93.09% over CAMELYON16 dataset. And the AUC over the cancer subtypes classification can be up to 96.03% and 98.82% over TCGA-NSCLC dataset and TCGA-RCC dataset, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge