Sourya Sengupta
Synthetic Volumetric Data Generation Enables Zero-Shot Generalization of Foundation Models in 3D Medical Image Segmentation
Jan 18, 2026Abstract:Foundation models such as Segment Anything Model 2 (SAM 2) exhibit strong generalization on natural images and videos but perform poorly on medical data due to differences in appearance statistics, imaging physics, and three-dimensional structure. To address this gap, we introduce SynthFM-3D, an analytical framework that mathematically models 3D variability in anatomy, contrast, boundary definition, and noise to generate synthetic data for training promptable segmentation models without real annotations. We fine-tuned SAM 2 on 10,000 SynthFM-3D volumes and evaluated it on eleven anatomical structures across three medical imaging modalities (CT, MR, ultrasound) from five public datasets. SynthFM-3D training led to consistent and statistically significant Dice score improvements over the pretrained SAM 2 baseline, demonstrating stronger zero-shot generalization across modalities. When compared with the supervised SAM-Med3D model on unseen cardiac ultrasound data, SynthFM-3D achieved 2-3x higher Dice scores, establishing analytical 3D data modeling as an effective pathway to modality-agnostic medical segmentation.
On the Utility of Virtual Staining for Downstream Applications as it relates to Task Network Capacity
Jul 31, 2025Abstract:Virtual staining, or in-silico-labeling, has been proposed to computationally generate synthetic fluorescence images from label-free images by use of deep learning-based image-to-image translation networks. In most reported studies, virtually stained images have been assessed only using traditional image quality measures such as structural similarity or signal-to-noise ratio. However, in biomedical imaging, images are typically acquired to facilitate an image-based inference, which we refer to as a downstream biological or clinical task. This study systematically investigates the utility of virtual staining for facilitating clinically relevant downstream tasks (like segmentation or classification) with consideration of the capacity of the deep neural networks employed to perform the tasks. Comprehensive empirical evaluations were conducted using biological datasets, assessing task performance by use of label-free, virtually stained, and ground truth fluorescence images. The results demonstrated that the utility of virtual staining is largely dependent on the ability of the segmentation or classification task network to extract meaningful task-relevant information, which is related to the concept of network capacity. Examples are provided in which virtual staining does not improve, or even degrades, segmentation or classification performance when the capacity of the associated task network is sufficiently large. The results demonstrate that task network capacity should be considered when deciding whether to perform virtual staining.
SynthFM: Training Modality-agnostic Foundation Models for Medical Image Segmentation without Real Medical Data
Apr 11, 2025



Abstract:Foundation models like the Segment Anything Model (SAM) excel in zero-shot segmentation for natural images but struggle with medical image segmentation due to differences in texture, contrast, and noise. Annotating medical images is costly and requires domain expertise, limiting large-scale annotated data availability. To address this, we propose SynthFM, a synthetic data generation framework that mimics the complexities of medical images, enabling foundation models to adapt without real medical data. Using SAM's pretrained encoder and training the decoder from scratch on SynthFM's dataset, we evaluated our method on 11 anatomical structures across 9 datasets (CT, MRI, and Ultrasound). SynthFM outperformed zero-shot baselines like SAM and MedSAM, achieving superior results under different prompt settings and on out-of-distribution datasets.
Is SAM 2 Better than SAM in Medical Image Segmentation?
Aug 08, 2024Abstract:Segment Anything Model (SAM) demonstrated impressive performance in zero-shot promptable segmentation on natural images. The recently released Segment Anything Model 2 (SAM 2) model claims to have better performance than SAM on images while extending the model's capabilities to video segmentation. It is important to evaluate the recent model's ability in medical image segmentation in a zero-shot promptable manner. In this work, we performed extensive studies with multiple datasets from different imaging modalities to compare the performance between SAM and SAM 2. We used two point prompt strategies: (i) single positive prompt near the centroid of the target structure and (ii) additional positive prompts placed randomly within the target structure. The evaluation included 21 unique organ-modality combinations including abdominal structures, cardiac structures, and fetal head images acquired from publicly available MRI, CT, and Ultrasound datasets. The preliminary results, based on 2D images, indicate that while SAM 2 may perform slightly better in a few cases, but it does not in general surpass SAM for medical image segmentation. Especially when the contrast is lower like in CT, Ultrasound images, SAM 2 performs poorly than SAM. For MRI images, SAM 2 performs at par or better than SAM. Similar to SAM, SAM 2 also suffers from over-segmentation issue especially when the boundaries of the to-be-segmented organ is fuzzy in nature.
Semi-Supervised Semantic Segmentation of Cell Nuclei via Diffusion-based Large-Scale Pre-Training and Collaborative Learning
Aug 08, 2023



Abstract:Automated semantic segmentation of cell nuclei in microscopic images is crucial for disease diagnosis and tissue microenvironment analysis. Nonetheless, this task presents challenges due to the complexity and heterogeneity of cells. While supervised deep learning methods are promising, they necessitate large annotated datasets that are time-consuming and error-prone to acquire. Semi-supervised approaches could provide feasible alternatives to this issue. However, the limited annotated data may lead to subpar performance of semi-supervised methods, regardless of the abundance of unlabeled data. In this paper, we introduce a novel unsupervised pre-training-based semi-supervised framework for cell-nuclei segmentation. Our framework is comprised of three main components. Firstly, we pretrain a diffusion model on a large-scale unlabeled dataset. The diffusion model's explicit modeling capability facilitates the learning of semantic feature representation from the unlabeled data. Secondly, we achieve semantic feature aggregation using a transformer-based decoder, where the pretrained diffusion model acts as the feature extractor, enabling us to fully utilize the small amount of labeled data. Finally, we implement a collaborative learning framework between the diffusion-based segmentation model and a supervised segmentation model to further enhance segmentation performance. Experiments were conducted on four publicly available datasets to demonstrate significant improvements compared to competitive semi-supervised segmentation methods and supervised baselines. A series of out-of-distribution tests further confirmed the generality of our framework. Furthermore, thorough ablation experiments and visual analysis confirmed the superiority of our proposed method.
Revisiting model self-interpretability in a decision-theoretic way for binary medical image classification
Mar 13, 2023Abstract:Interpretability is highly desired for deep neural network-based classifiers, especially when addressing high-stake decisions in medical imaging. Commonly used post-hoc interpretability methods may not be always useful because different such methods can produce several plausible but different interpretations of a given model, leading to confusion about which one to choose. {In this work, an {inherently} interpretable encoder-decoder model coupled with a single-layer fully connected network with unity weights is proposed for binary medical image classification problems. The feature extraction component of a trained black-box network for the same task is employed as the pre-trained encoder of the interpretable model. The model is trained to estimate the decision statistic of the given trained black-box deep binary classifier to maintain a similar accuracy.} The decoder output represents a transformed version of the to-be-classified image that, when processed by the fixed fully connected layer, produces the same decision statistic value as the original classifier. This is accomplished by minimizing the mean squared error between the decision statistic values of the black-box model and encoder-decoder based model during training. The decoder output image is referred to as an equivalency map. Because the single-layer network is fully interpretable, the equivalency map provides a visualization of the transformed image features that contribute to the decision statistic value and, moreover, permits quantification of their relative contributions. Unlike the traditional post-hoc interpretability methods, the proposed method is inherently interpretable, quantitative, and fundamentally based on decision theory.
De-speckling of Optical Coherence Tomography Images Using Anscombe Transform and a Noisier2noise Model
Sep 20, 2022


Abstract:Optical Coherence Tomography (OCT) image denoising is a fundamental problem as OCT images suffer from multiplicative speckle noise, resulting in poor visibility of retinal layers. The traditional denoising methods consider specific statistical properties of the noise, which are not always known. Furthermore, recent deep learning-based denoising methods require paired noisy and clean images, which are often difficult to obtain, especially medical images. Noise2Noise family architectures are generally proposed to overcome this issue by learning without noisy-clean image pairs. However, for that, multiple noisy observations from a single image are typically needed. Also, sometimes the experiments are demonstrated by simulating noises on clean synthetic images, which is not a realistic scenario. This work shows how a single real-world noisy observation of each image can be used to train a denoising network. Along with a theoretical understanding, our algorithm is experimentally validated using a publicly available OCT image dataset. Our approach incorporates Anscombe transform to convert the multiplicative noise model to additive Gaussian noise to make it suitable for OCT images. The quantitative results show that this method can outperform several other methods where a single noisy observation of an image is needed for denoising. The code and implementation of this paper will be available publicly upon acceptance of this paper.
Uncertainty aware and explainable diagnosis of retinal disease
Jan 26, 2021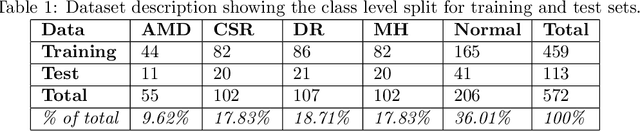
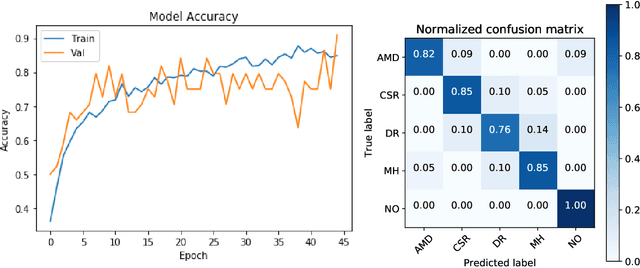
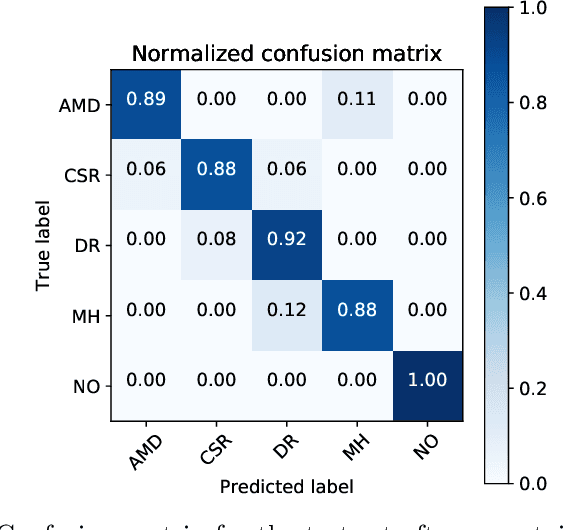
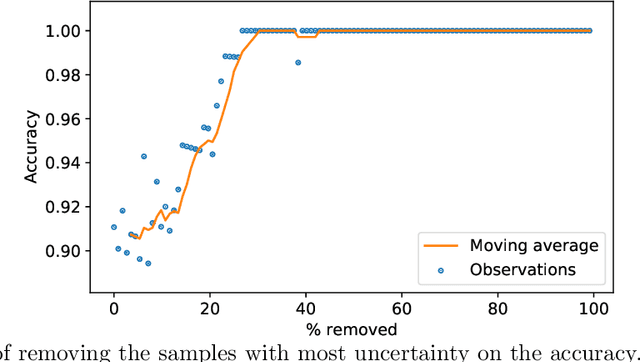
Abstract:Deep learning methods for ophthalmic diagnosis have shown considerable success in tasks like segmentation and classification. However, their widespread application is limited due to the models being opaque and vulnerable to making a wrong decision in complicated cases. Explainability methods show the features that a system used to make prediction while uncertainty awareness is the ability of a system to highlight when it is not sure about the decision. This is one of the first studies using uncertainty and explanations for informed clinical decision making. We perform uncertainty analysis of a deep learning model for diagnosis of four retinal diseases - age-related macular degeneration (AMD), central serous retinopathy (CSR), diabetic retinopathy (DR), and macular hole (MH) using images from a publicly available (OCTID) dataset. Monte Carlo (MC) dropout is used at the test time to generate a distribution of parameters and the predictions approximate the predictive posterior of a Bayesian model. A threshold is computed using the distribution and uncertain cases can be referred to the ophthalmologist thus avoiding an erroneous diagnosis. The features learned by the model are visualized using a proven attribution method from a previous study. The effects of uncertainty on model performance and the relationship between uncertainty and explainability are discussed in terms of clinical significance. The uncertainty information along with the heatmaps make the system more trustworthy for use in clinical settings.
Explainable deep learning models in medical image analysis
May 28, 2020



Abstract:Deep learning methods have been very effective for a variety of medical diagnostic tasks and has even beaten human experts on some of those. However, the black-box nature of the algorithms has restricted clinical use. Recent explainability studies aim to show the features that influence the decision of a model the most. The majority of literature reviews of this area have focused on taxonomy, ethics, and the need for explanations. A review of the current applications of explainable deep learning for different medical imaging tasks is presented here. The various approaches, challenges for clinical deployment, and the areas requiring further research are discussed here from a practical standpoint of a deep learning researcher designing a system for the clinical end-users.
Ophthalmic Diagnosis and Deep Learning -- A Survey
Dec 09, 2018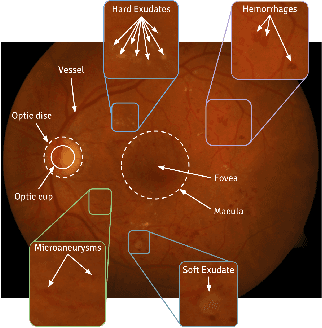
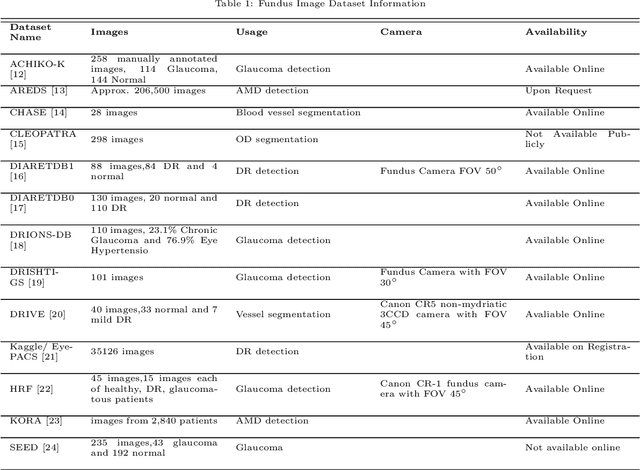
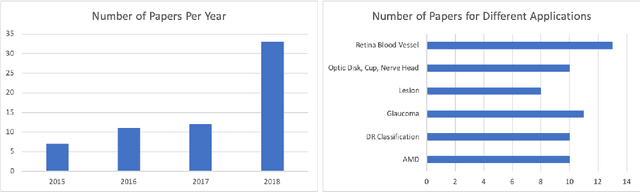
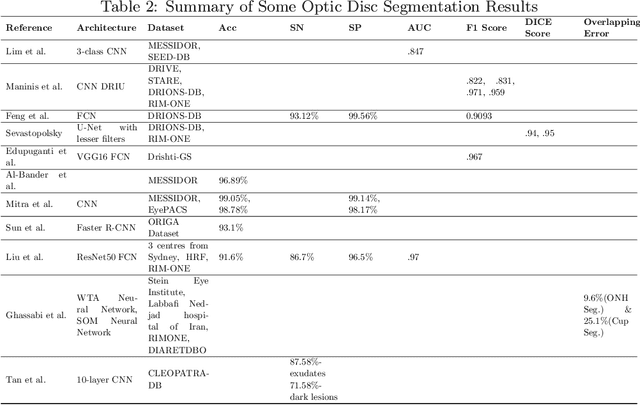
Abstract:This survey paper presents a detailed overview of the applications for deep learning in ophthalmic diagnosis using retinal imaging techniques. The need of automated computer-aided deep learning models for medical diagnosis is discussed. Then a detailed review of the available retinal image datasets is provided. Applications of deep learning for segmentation of optic disk, blood vessels and retinal layer as well as detection of red lesions are reviewed.Recent deep learning models for classification of retinal disease including age-related macular degeneration, glaucoma, diabetic macular edema and diabetic retinopathy are also reported.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge