Jinfeng Wang
Guangdong Power Grid Co.
One-Shot Price Forecasting with Covariate-Guided Experts under Privacy Constraints
Jan 17, 2026Abstract:Forecasting in power systems often involves multivariate time series with complex dependencies and strict privacy constraints across regions. Traditional forecasting methods require significant expert knowledge and struggle to generalize across diverse deployment scenarios. Recent advancements in pre-trained time series models offer new opportunities, but their zero-shot performance on domain-specific tasks remains limited. To address these challenges, we propose a novel MoE Encoder module that augments pretrained forecasting models by injecting a sparse mixture-of-experts layer between tokenization and encoding. This design enables two key capabilities: (1) trans forming multivariate forecasting into an expert-guided univariate task, allowing the model to effectively capture inter-variable relations, and (2) supporting localized training and lightweight parameter sharing in federated settings where raw data cannot be exchanged. Extensive experiments on public multivariate datasets demonstrate that MoE-Encoder significantly improves forecasting accuracy compared to strong baselines. We further simulate federated environments and show that transferring only MoE-Encoder parameters allows efficient adaptation to new regions, with minimal performance degradation. Our findings suggest that MoE-Encoder provides a scalable and privacy-aware extension to foundation time series models.
Contrastive Learning Via Equivariant Representation
Jun 01, 2024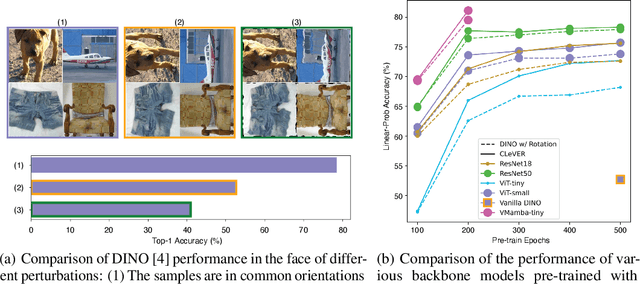
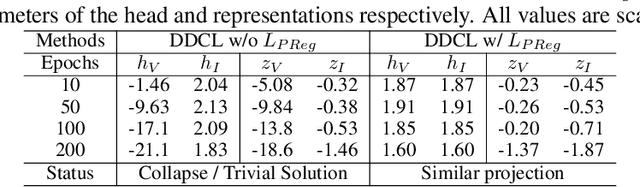
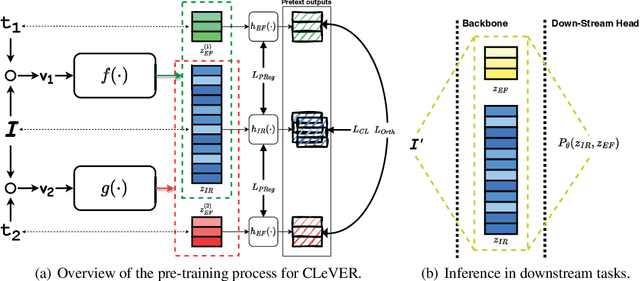
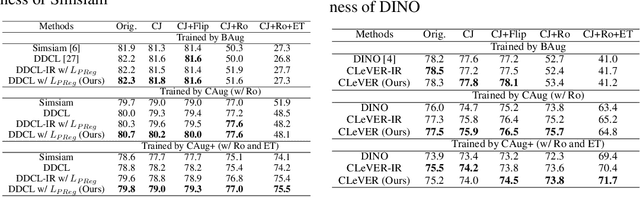
Abstract:Invariant-based Contrastive Learning (ICL) methods have achieved impressive performance across various domains. However, the absence of latent space representation for distortion (augmentation)-related information in the latent space makes ICL sub-optimal regarding training efficiency and robustness in downstream tasks. Recent studies suggest that introducing equivariance into Contrastive Learning (CL) can improve overall performance. In this paper, we rethink the roles of augmentation strategies and equivariance in improving CL efficacy. We propose a novel Equivariant-based Contrastive Learning (ECL) framework, CLeVER (Contrastive Learning Via Equivariant Representation), compatible with augmentation strategies of arbitrary complexity for various mainstream CL methods and model frameworks. Experimental results demonstrate that CLeVER effectively extracts and incorporates equivariant information from data, thereby improving the training efficiency and robustness of baseline models in downstream tasks.
ProMISe: Promptable Medical Image Segmentation using SAM
Mar 07, 2024



Abstract:With the proposal of the Segment Anything Model (SAM), fine-tuning SAM for medical image segmentation (MIS) has become popular. However, due to the large size of the SAM model and the significant domain gap between natural and medical images, fine-tuning-based strategies are costly with potential risk of instability, feature damage and catastrophic forgetting. Furthermore, some methods of transferring SAM to a domain-specific MIS through fine-tuning strategies disable the model's prompting capability, severely limiting its utilization scenarios. In this paper, we propose an Auto-Prompting Module (APM), which provides SAM-based foundation model with Euclidean adaptive prompts in the target domain. Our experiments demonstrate that such adaptive prompts significantly improve SAM's non-fine-tuned performance in MIS. In addition, we propose a novel non-invasive method called Incremental Pattern Shifting (IPS) to adapt SAM to specific medical domains. Experimental results show that the IPS enables SAM to achieve state-of-the-art or competitive performance in MIS without the need for fine-tuning. By coupling these two methods, we propose ProMISe, an end-to-end non-fine-tuned framework for Promptable Medical Image Segmentation. Our experiments demonstrate that both using our methods individually or in combination achieves satisfactory performance in low-cost pattern shifting, with all of SAM's parameters frozen.
Atrial Septal Defect Detection in Children Based on Ultrasound Video Using Multiple Instances Learning
Jun 06, 2023

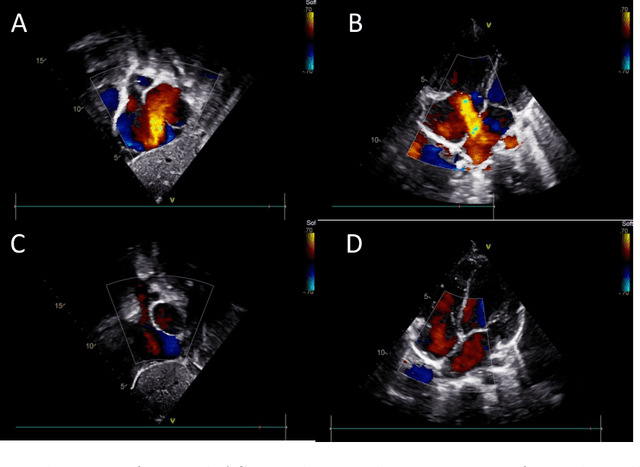
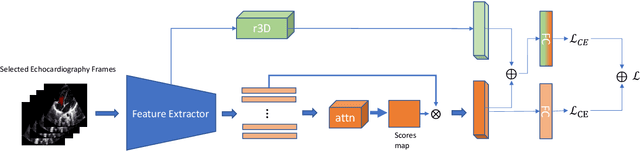
Abstract:Purpose: Congenital heart defect (CHD) is the most common birth defect. Thoracic echocardiography (TTE) can provide sufficient cardiac structure information, evaluate hemodynamics and cardiac function, and is an effective method for atrial septal defect (ASD) examination. This paper aims to study a deep learning method based on cardiac ultrasound video to assist in ASD diagnosis. Materials and methods: We select two standard views of the atrial septum (subAS) and low parasternal four-compartment view (LPS4C) as the two views to identify ASD. We enlist data from 300 children patients as part of a double-blind experiment for five-fold cross-validation to verify the performance of our model. In addition, data from 30 children patients (15 positives and 15 negatives) are collected for clinician testing and compared to our model test results (these 30 samples do not participate in model training). We propose an echocardiography video-based atrial septal defect diagnosis system. In our model, we present a block random selection, maximal agreement decision and frame sampling strategy for training and testing respectively, resNet18 and r3D networks are used to extract the frame features and aggregate them to build a rich video-level representation. Results: We validate our model using our private dataset by five-cross validation. For ASD detection, we achieve 89.33 AUC, 84.95 accuracy, 85.70 sensitivity, 81.51 specificity and 81.99 F1 score. Conclusion: The proposed model is multiple instances learning-based deep learning model for video atrial septal defect detection which effectively improves ASD detection accuracy when compared to the performances of previous networks and clinical doctors.
Distortion-Disentangled Contrastive Learning
Mar 09, 2023



Abstract:Self-supervised learning is well known for its remarkable performance in representation learning and various downstream computer vision tasks. Recently, Positive-pair-Only Contrastive Learning (POCL) has achieved reliable performance without the need to construct positive-negative training sets. It reduces memory requirements by lessening the dependency on the batch size. The POCL method typically uses a single loss function to extract the distortion invariant representation (DIR) which describes the proximity of positive-pair representations affected by different distortions. This loss function implicitly enables the model to filter out or ignore the distortion variant representation (DVR) affected by different distortions. However, existing POCL methods do not explicitly enforce the disentanglement and exploitation of the actually valuable DVR. In addition, these POCL methods have been observed to be sensitive to augmentation strategies. To address these limitations, we propose a novel POCL framework named Distortion-Disentangled Contrastive Learning (DDCL) and a Distortion-Disentangled Loss (DDL). Our approach is the first to explicitly disentangle and exploit the DVR inside the model and feature stream to improve the overall representation utilization efficiency, robustness and representation ability. Experiments carried out demonstrate the superiority of our framework to Barlow Twins and Simsiam in terms of convergence, representation quality, and robustness on several benchmark datasets.
Bilateral-Fuser: A Novel Multi-cue Fusion Architecture with Anatomical-aware Tokens for Fovea Localization
Feb 14, 2023



Abstract:Accurate localization of fovea is one of the primary steps in analyzing retinal diseases since it helps prevent irreversible vision loss. Although current deep learning-based methods achieve better performance than traditional methods, there still remain challenges such as utilizing anatomical landmarks insufficiently, sensitivity to diseased retinal images and various image conditions. In this paper, we propose a novel transformer-based architecture (Bilateral-Fuser) for multi-cue fusion. This architecture explicitly incorporates long-range connections and global features using retina and vessel distributions for robust fovea localization. We introduce a spatial attention mechanism in the dual-stream encoder for extracting and fusing self-learned anatomical information. This design focuses more on features distributed along blood vessels and significantly decreases computational costs by reducing token numbers. Our comprehensive experiments show that the proposed architecture achieves state-of-the-art performance on two public and one large-scale private datasets. We also present that the Bilateral-Fuser is more robust on both normal and diseased retina images and has better generalization capacity in cross-dataset experiments.
DuAT: Dual-Aggregation Transformer Network for Medical Image Segmentation
Dec 21, 2022



Abstract:Transformer-based models have been widely demonstrated to be successful in computer vision tasks by modelling long-range dependencies and capturing global representations. However, they are often dominated by features of large patterns leading to the loss of local details (e.g., boundaries and small objects), which are critical in medical image segmentation. To alleviate this problem, we propose a Dual-Aggregation Transformer Network called DuAT, which is characterized by two innovative designs, namely, the Global-to-Local Spatial Aggregation (GLSA) and Selective Boundary Aggregation (SBA) modules. The GLSA has the ability to aggregate and represent both global and local spatial features, which are beneficial for locating large and small objects, respectively. The SBA module is used to aggregate the boundary characteristic from low-level features and semantic information from high-level features for better preserving boundary details and locating the re-calibration objects. Extensive experiments in six benchmark datasets demonstrate that our proposed model outperforms state-of-the-art methods in the segmentation of skin lesion images, and polyps in colonoscopy images. In addition, our approach is more robust than existing methods in various challenging situations such as small object segmentation and ambiguous object boundaries.
Stepwise Feature Fusion: Local Guides Global
Mar 07, 2022



Abstract:Colonoscopy, currently the most efficient and recognized colon polyp detection technology, is necessary for early screening and prevention of colorectal cancer. However, due to the varying size and complex morphological features of colonic polyps as well as the indistinct boundary between polyps and mucosa, accurate segmentation of polyps is still challenging. Deep learning has become popular for accurate polyp segmentation tasks with excellent results. However, due to the structure of polyps image and the varying shapes of polyps, it easy for existing deep learning models to overfitting the current dataset. As a result, the model may not process unseen colonoscopy data. To address this, we propose a new State-Of-The-Art model for medical image segmentation, the SSFormer, which uses a pyramid Transformer encoder to improve the generalization ability of models. Specifically, our proposed Progressive Locality Decoder can be adapted to the pyramid Transformer backbone to emphasize local features and restrict attention dispersion. The SSFormer achieves statet-of-the-art performance in both learning and generalization assessment.
A Robust Framework of Chromosome Straightening with ViT-Patch GAN
Mar 06, 2022



Abstract:Chromosomes exhibit non-rigid and non-articulated nature with varying degrees of curvature. Chromosome straightening is an essential step for subsequent karyotype construction, pathological diagnosis and cytogenetic map development. However, robust chromosome straightening remains challenging, due to the unavailability of training images, distorted chromosome details and shapes after straightening, as well as poor generalization capability. We propose a novel architecture, ViT-Patch GAN, consisting of a motion transformation generator and a Vision Transformer-based patch (ViT-Patch) discriminator. The generator learns the motion representation of chromosomes for straightening. With the help of the ViT-Patch discriminator, the straightened chromosomes retain more shape and banding pattern details. The proposed framework is trained on a small dataset and is able to straighten chromosome images with state-of-the-art performance for two large datasets.
Autoencoder Based Residual Deep Networks for Robust Regression Prediction and Spatiotemporal Estimation
Dec 29, 2018



Abstract:To have a superior generalization, a deep learning neural network often involves a large size of training sample. With increase of hidden layers in order to increase learning ability, neural network has potential degradation in accuracy. Both could seriously limit applicability of deep learning in some domains particularly involving predictions of continuous variables with a small size of samples. Inspired by residual convolutional neural network in computer vision and recent findings of crucial shortcuts in the brains in neuroscience, we propose an autoencoder-based residual deep network for robust prediction. In a nested way, we leverage shortcut connections to implement residual mapping with a balanced structure for efficient propagation of error signals. The novel method is demonstrated by multiple datasets, imputation of high spatiotemporal resolution non-randomness missing values of aerosol optical depth, and spatiotemporal estimation of fine particulate matter <2.5 \mu m, achieving the cutting edge of accuracy and efficiency. Our approach is also a general-purpose regression learner to be applicable in diverse domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge