Boxiang Yun
MDN: Mamba-Driven Dualstream Network For Medical Hyperspectral Image Segmentation
Feb 24, 2025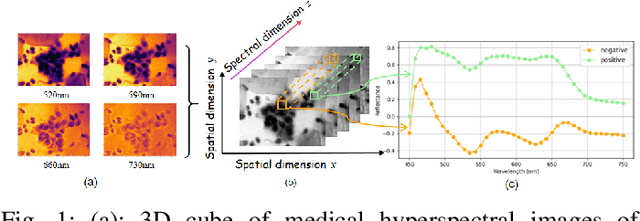
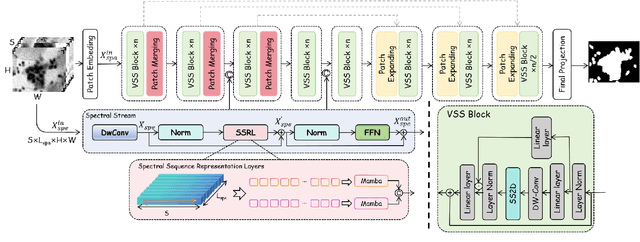
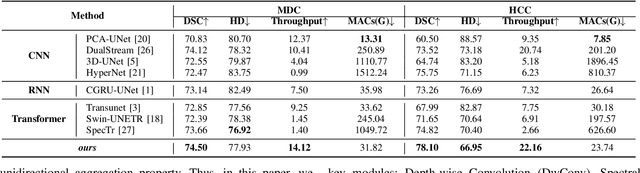
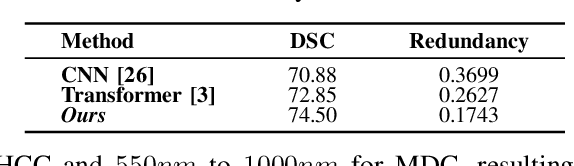
Abstract:Medical Hyperspectral Imaging (MHSI) offers potential for computational pathology and precision medicine. However, existing CNN and Transformer struggle to balance segmentation accuracy and speed due to high spatial-spectral dimensionality. In this study, we leverage Mamba's global context modeling to propose a dual-stream architecture for joint spatial-spectral feature extraction. To address the limitation of Mamba's unidirectional aggregation, we introduce a recurrent spectral sequence representation to capture low-redundancy global spectral features. Experiments on a public Multi-Dimensional Choledoch dataset and a private Cervical Cancer dataset show that our method outperforms state-of-the-art approaches in segmentation accuracy while minimizing resource usage and achieving the fastest inference speed. Our code will be available at https://github.com/DeepMed-Lab-ECNU/MDN.
FS-MedSAM2: Exploring the Potential of SAM2 for Few-Shot Medical Image Segmentation without Fine-tuning
Sep 06, 2024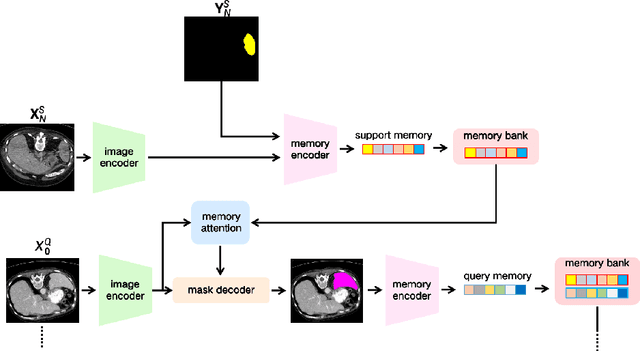


Abstract:The Segment Anything Model 2 (SAM2) has recently demonstrated exceptional performance in zero-shot prompt segmentation for natural images and videos. However, it faces significant challenges when applied to medical images. Since its release, many attempts have been made to adapt SAM2's segmentation capabilities to the medical imaging domain. These efforts typically involve using a substantial amount of labeled data to fine-tune the model's weights. In this paper, we explore SAM2 from a different perspective via making the full use of its trained memory attention module and its ability of processing mask prompts. We introduce FS-MedSAM2, a simple yet effective framework that enables SAM2 to achieve superior medical image segmentation in a few-shot setting, without the need for fine-tuning. Our framework outperforms the current state-of-the-arts on two publicly available medical image datasets. The code is available at https://github.com/DeepMed-Lab-ECNU/FS_MedSAM2.
Boosting Open-Domain Continual Learning via Leveraging Intra-domain Category-aware Prototype
Aug 19, 2024Abstract:Despite recent progress in enhancing the efficacy of Open-Domain Continual Learning (ODCL) in Vision-Language Models (VLM), failing to (1) correctly identify the Task-ID of a test image and (2) use only the category set corresponding to the Task-ID, while preserving the knowledge related to each domain, cannot address the two primary challenges of ODCL: forgetting old knowledge and maintaining zero-shot capabilities, as well as the confusions caused by category-relatedness between domains. In this paper, we propose a simple yet effective solution: leveraging intra-domain category-aware prototypes for ODCL in CLIP (DPeCLIP), where the prototype is the key to bridging the above two processes. Concretely, we propose a training-free Task-ID discriminator method, by utilizing prototypes as classifiers for identifying Task-IDs. Furthermore, to maintain the knowledge corresponding to each domain, we incorporate intra-domain category-aware prototypes as domain prior prompts into the training process. Extensive experiments conducted on 11 different datasets demonstrate the effectiveness of our approach, achieving 2.37% and 1.14% average improvement in class-incremental and task-incremental settings, respectively.
SpecTr: Spectral Transformer for Hyperspectral Pathology Image Segmentation
Mar 05, 2021



Abstract:Hyperspectral imaging (HSI) unlocks the huge potential to a wide variety of applications relied on high-precision pathology image segmentation, such as computational pathology and precision medicine. Since hyperspectral pathology images benefit from the rich and detailed spectral information even beyond the visible spectrum, the key to achieve high-precision hyperspectral pathology image segmentation is to felicitously model the context along high-dimensional spectral bands. Inspired by the strong context modeling ability of transformers, we hereby, for the first time, formulate the contextual feature learning across spectral bands for hyperspectral pathology image segmentation as a sequence-to-sequence prediction procedure by transformers. To assist spectral context learning procedure, we introduce two important strategies: (1) a sparsity scheme enforces the learned contextual relationship to be sparse, so as to eliminates the distraction from the redundant bands; (2) a spectral normalization, a separate group normalization for each spectral band, mitigates the nuisance caused by heterogeneous underlying distributions of bands. We name our method Spectral Transformer (SpecTr), which enjoys two benefits: (1) it has a strong ability to model long-range dependency among spectral bands, and (2) it jointly explores the spatial-spectral features of HSI. Experiments show that SpecTr outperforms other competing methods in a hyperspectral pathology image segmentation benchmark without the need of pre-training. Code is available at https://github.com/hfut-xc-yun/SpecTr.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge