Wanxing Chang
From Histopathology Images to Cell Clouds: Learning Slide Representations with Hierarchical Cell Transformer
Dec 21, 2024Abstract:It is clinically crucial and potentially very beneficial to be able to analyze and model directly the spatial distributions of cells in histopathology whole slide images (WSI). However, most existing WSI datasets lack cell-level annotations, owing to the extremely high cost over giga-pixel images. Thus, it remains an open question whether deep learning models can directly and effectively analyze WSIs from the semantic aspect of cell distributions. In this work, we construct a large-scale WSI dataset with more than 5 billion cell-level annotations, termed WSI-Cell5B, and a novel hierarchical Cell Cloud Transformer (CCFormer) to tackle these challenges. WSI-Cell5B is based on 6,998 WSIs of 11 cancers from The Cancer Genome Atlas Program, and all WSIs are annotated per cell by coordinates and types. To the best of our knowledge, WSI-Cell5B is the first WSI-level large-scale dataset integrating cell-level annotations. On the other hand, CCFormer formulates the collection of cells in each WSI as a cell cloud and models cell spatial distribution. Specifically, Neighboring Information Embedding (NIE) is proposed to characterize the distribution of cells within the neighborhood of each cell, and a novel Hierarchical Spatial Perception (HSP) module is proposed to learn the spatial relationship among cells in a bottom-up manner. The clinical analysis indicates that WSI-Cell5B can be used to design clinical evaluation metrics based on counting cells that effectively assess the survival risk of patients. Extensive experiments on survival prediction and cancer staging show that learning from cell spatial distribution alone can already achieve state-of-the-art (SOTA) performance, i.e., CCFormer strongly outperforms other competing methods.
From Pixels to Gigapixels: Bridging Local Inductive Bias and Long-Range Dependencies with Pixel-Mamba
Dec 21, 2024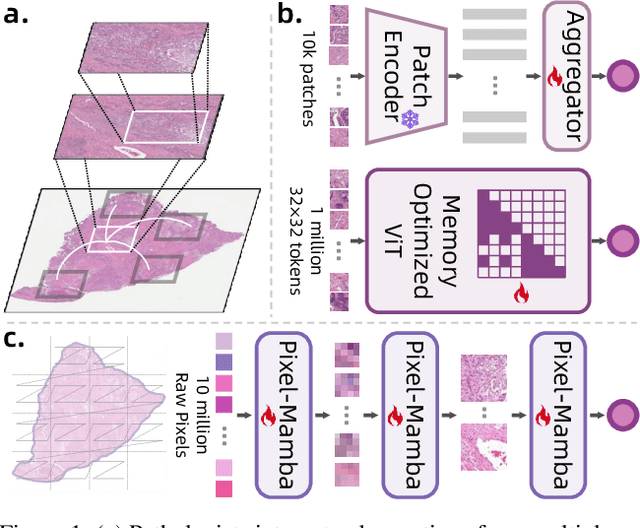
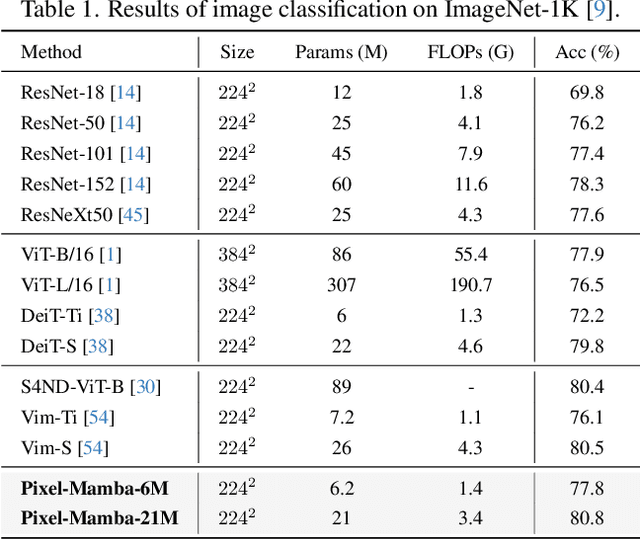
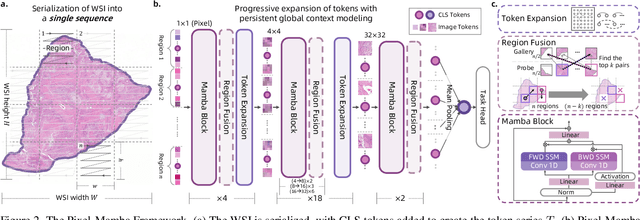
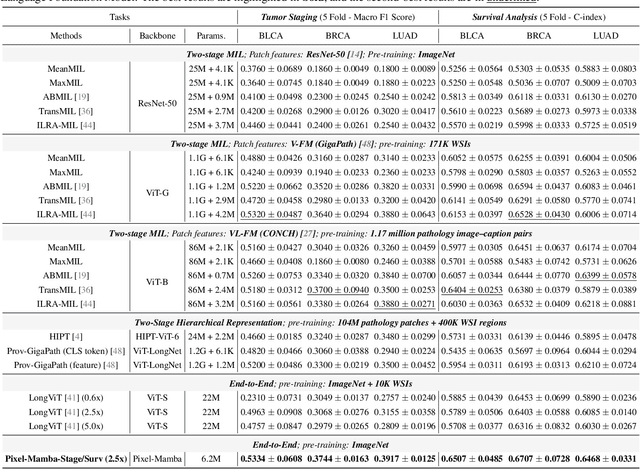
Abstract:Histopathology plays a critical role in medical diagnostics, with whole slide images (WSIs) offering valuable insights that directly influence clinical decision-making. However, the large size and complexity of WSIs may pose significant challenges for deep learning models, in both computational efficiency and effective representation learning. In this work, we introduce Pixel-Mamba, a novel deep learning architecture designed to efficiently handle gigapixel WSIs. Pixel-Mamba leverages the Mamba module, a state-space model (SSM) with linear memory complexity, and incorporates local inductive biases through progressively expanding tokens, akin to convolutional neural networks. This enables Pixel-Mamba to hierarchically combine both local and global information while efficiently addressing computational challenges. Remarkably, Pixel-Mamba achieves or even surpasses the quantitative performance of state-of-the-art (SOTA) foundation models that were pretrained on millions of WSIs or WSI-text pairs, in a range of tumor staging and survival analysis tasks, {\bf even without requiring any pathology-specific pretraining}. Extensive experiments demonstrate the efficacy of Pixel-Mamba as a powerful and efficient framework for end-to-end WSI analysis.
CSOT: Curriculum and Structure-Aware Optimal Transport for Learning with Noisy Labels
Dec 11, 2023



Abstract:Learning with noisy labels (LNL) poses a significant challenge in training a well-generalized model while avoiding overfitting to corrupted labels. Recent advances have achieved impressive performance by identifying clean labels and correcting corrupted labels for training. However, the current approaches rely heavily on the model's predictions and evaluate each sample independently without considering either the global and local structure of the sample distribution. These limitations typically result in a suboptimal solution for the identification and correction processes, which eventually leads to models overfitting to incorrect labels. In this paper, we propose a novel optimal transport (OT) formulation, called Curriculum and Structure-aware Optimal Transport (CSOT). CSOT concurrently considers the inter- and intra-distribution structure of the samples to construct a robust denoising and relabeling allocator. During the training process, the allocator incrementally assigns reliable labels to a fraction of the samples with the highest confidence. These labels have both global discriminability and local coherence. Notably, CSOT is a new OT formulation with a nonconvex objective function and curriculum constraints, so it is not directly compatible with classical OT solvers. Here, we develop a lightspeed computational method that involves a scaling iteration within a generalized conditional gradient framework to solve CSOT efficiently. Extensive experiments demonstrate the superiority of our method over the current state-of-the-arts in LNL. Code is available at https://github.com/changwxx/CSOT-for-LNL.
Unified Optimal Transport Framework for Universal Domain Adaptation
Oct 31, 2022Abstract:Universal Domain Adaptation (UniDA) aims to transfer knowledge from a source domain to a target domain without any constraints on label sets. Since both domains may hold private classes, identifying target common samples for domain alignment is an essential issue in UniDA. Most existing methods require manually specified or hand-tuned threshold values to detect common samples thus they are hard to extend to more realistic UniDA because of the diverse ratios of common classes. Moreover, they cannot recognize different categories among target-private samples as these private samples are treated as a whole. In this paper, we propose to use Optimal Transport (OT) to handle these issues under a unified framework, namely UniOT. First, an OT-based partial alignment with adaptive filling is designed to detect common classes without any predefined threshold values for realistic UniDA. It can automatically discover the intrinsic difference between common and private classes based on the statistical information of the assignment matrix obtained from OT. Second, we propose an OT-based target representation learning that encourages both global discrimination and local consistency of samples to avoid the over-reliance on the source. Notably, UniOT is the first method with the capability to automatically discover and recognize private categories in the target domain for UniDA. Accordingly, we introduce a new metric H^3-score to evaluate the performance in terms of both accuracy of common samples and clustering performance of private ones. Extensive experiments clearly demonstrate the advantages of UniOT over a wide range of state-of-the-art methods in UniDA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge